Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1
- PMID: 17237364
- PMCID: PMC1828933
- DOI: 10.1128/EC.00264-06
Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1
Abstract
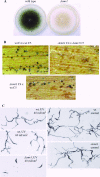
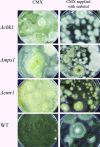
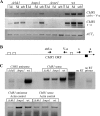
The maize pathogen Cochliobolus heterostrophus requires two mitogen-activated protein kinases (MAPKs), Chk1 and Mps1, to produce normal pigmentation. Young colonies of mps1 and chk1 deletion mutants have a white and autolytic appearance, which was partially rescued by a hyperosmotic environment. We isolated the transcription factor Cmr1, an ortholog of Colletotrichum lagenarium Cmr1 and Magnaporthe grisea Pig1, which regulates melanin biosynthesis in C. heterostrophus. Deletion of CMR1 in C. heterostrophus resulted in mutants that lacked dark pigmentation and acquired an orange-pink color. In cmr1 deletion strains the expression of putative scytalone dehydratase (SCD1) and hydroxynaphthalene reductase (BRN1 and BRN2) genes involved in melanin biosynthesis was undetectable, whereas expression of PKS18, encoding a polyketide synthase, was only moderately reduced. In chk1 and mps1 mutants expression of PKS18, SCD1, BRN1, BRN2, and the transcription factor CMR1 itself was very low in young colonies, slightly up-regulated in aging colonies, and significantly induced in hyperosmotic conditions, compared to invariably high expression in the wild type. These findings indicate that two MAPKs, Chk1 and Mps1, affect Cmr1 at the transcriptional level and this influence is partially overridden in stress conditions including aging culture and hyperosmotic environment. Surprisingly, we found that the CMR1 gene was transcribed in both sense and antisense directions, apparently producing mRNA as well as a long noncoding RNA transcript. Expression of the antisense CMR1 was also Chk1 and Mps1 dependent. Analysis of chromosomal location of the melanin biosynthesis genes in C. heterostrophus resulted in identification of a small gene cluster comprising BRN1, CMR1, and PKS18. Since expression of all three genes depends on Chk1 and Mps1 MAPKs, we suggest their possible epigenetic regulation.
Figures
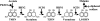




References
-
- Brakhage, A. A., and B. Liebmann. 2005. Aspergillus fumigatus conidial pigment and cAMP signal transduction: significance for virulence. Med. Mycol. 43(Suppl. 1):S75-S82. - PubMed
-
- Braun, E., and R. Howard. 1994. Adhesion of Cochliobolus heterostrophus conidia and germlings to leaves and artificial surfaces. Exp. Mycol. 18:211-220.
-
- Di Pietro, A., F. I. Garcia-MacEira, E. Meglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. - PubMed
-
- Guillen, A., B. G. Turgeon, P. R. Thorson, C. R. Bronson, and O. C. Yoder. 1994. Linkage among melanin biosynthetic mutations in Cochliobolus heterostrophus. Fungal Genet. Newsl. 41:40.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

