Inference of population structure under a Dirichlet process model
- PMID: 17237522
- PMCID: PMC1855109
- DOI: 10.1534/genetics.106.061317
Inference of population structure under a Dirichlet process model
Abstract
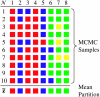
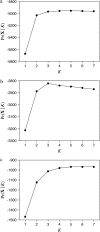
Inferring population structure from genetic data sampled from some number of individuals is a formidable statistical problem. One widely used approach considers the number of populations to be fixed and calculates the posterior probability of assigning individuals to each population. More recently, the assignment of individuals to populations and the number of populations have both been considered random variables that follow a Dirichlet process prior. We examined the statistical behavior of assignment of individuals to populations under a Dirichlet process prior. First, we examined a best-case scenario, in which all of the assumptions of the Dirichlet process prior were satisfied, by generating data under a Dirichlet process prior. Second, we examined the performance of the method when the genetic data were generated under a population genetics model with symmetric migration between populations. We examined the accuracy of population assignment using a distance on partitions. The method can be quite accurate with a moderate number of loci. As expected, inferences on the number of populations are more accurate when theta = 4N(e)u is large and when the migration rate (4N(e)m) is low. We also examined the sensitivity of inferences of population structure to choice of the parameter of the Dirichlet process model. Although inferences could be sensitive to the choice of the prior on the number of populations, this sensitivity occurred when the number of loci sampled was small; inferences are more robust to the prior on the number of populations when the number of sampled loci is large. Finally, we discuss several methods for summarizing the results of a Bayesian Markov chain Monte Carlo (MCMC) analysis of population structure. We develop the notion of the mean population partition, which is the partition of individuals to populations that minimizes the squared partition distance to the partitions sampled by the MCMC algorithm.
Figures




Similar articles
-
Structurama: bayesian inference of population structure.Evol Bioinform Online. 2011;7:55-9. doi: 10.4137/EBO.S6761. Epub 2011 Jun 2. Evol Bioinform Online. 2011. PMID: 21698091 Free PMC article.
-
Characterization of a Bayesian genetic clustering algorithm based on a Dirichlet process prior and comparison among Bayesian clustering methods.BMC Bioinformatics. 2011 Jun 28;12:263. doi: 10.1186/1471-2105-12-263. BMC Bioinformatics. 2011. PMID: 21708038 Free PMC article.
-
Mixed model analysis of quantitative trait loci.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14542-7. doi: 10.1073/pnas.250235197. Proc Natl Acad Sci U S A. 2000. PMID: 11114174 Free PMC article.
-
Estimating genealogies from unlinked marker data: a Bayesian approach.Theor Popul Biol. 2007 Nov;72(3):305-22. doi: 10.1016/j.tpb.2007.06.004. Epub 2007 Jun 22. Theor Popul Biol. 2007. PMID: 17681576 Review.
-
A Markov chain Monte Carlo strategy for sampling from the joint posterior distribution of pedigrees and population parameters under a Fisher-Wright model with partial selfing.Theor Popul Biol. 2007 Nov;72(3):436-58. doi: 10.1016/j.tpb.2007.03.002. Epub 2007 Mar 12. Theor Popul Biol. 2007. PMID: 17448511 Review.
Cited by
-
Estimating the Number of Subpopulations (K) in Structured Populations.Genetics. 2016 Aug;203(4):1827-39. doi: 10.1534/genetics.115.180992. Epub 2016 Jun 17. Genetics. 2016. PMID: 27317680 Free PMC article.
-
Landscape genetics reveals focal transmission of a human macroparasite.PLoS Negl Trop Dis. 2010 Apr 20;4(4):e665. doi: 10.1371/journal.pntd.0000665. PLoS Negl Trop Dis. 2010. PMID: 20421919 Free PMC article.
-
Empirical Bayes inference of pairwise F(ST) and its distribution in the genome.Genetics. 2007 Oct;177(2):861-73. doi: 10.1534/genetics.107.077263. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660541 Free PMC article.
-
Variations on a common STRUCTURE: new algorithms for a valuable model.Genetics. 2014 Jul;197(3):809-11. doi: 10.1534/genetics.114.166264. Genetics. 2014. PMID: 25024035 Free PMC article. No abstract available.
-
Unravelling the hidden ancestry of American admixed populations.Nat Commun. 2015 Mar 24;6:6596. doi: 10.1038/ncomms7596. Nat Commun. 2015. PMID: 25803618 Free PMC article.
References
-
- Akaike, H., 1973. Information theory as an extension of the maximum likelihood principle, pp. 267–281 in Second International Symposium on Information Theory, edited by B. N. Petrov and F. Csaki. Akademiai Kiado, Budapest.
-
- Antoniak, C. E., 1974. Mixtures of Dirichlet processes with applications to non-parametric problems. Ann. Stat. 2: 1152–1174.
-
- Balding, D. J., and R. A. Nichols, 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. - PubMed
-
- Bell, E. T., 1934. Exponential numbers. Am. Math. Mon. 41: 411–419.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

