Foxp3 occupancy and regulation of key target genes during T-cell stimulation
- PMID: 17237765
- PMCID: PMC3008159
- DOI: 10.1038/nature05478
Foxp3 occupancy and regulation of key target genes during T-cell stimulation
Abstract
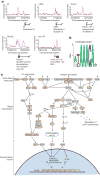
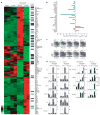
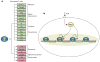
Foxp3+CD4+CD25+ regulatory T (T(reg)) cells are essential for the prevention of autoimmunity. T(reg) cells have an attenuated cytokine response to T-cell receptor stimulation, and can suppress the proliferation and effector function of neighbouring T cells. The forkhead transcription factor Foxp3 (forkhead box P3) is selectively expressed in T(reg) cells, is required for T(reg) development and function, and is sufficient to induce a T(reg) phenotype in conventional CD4+CD25- T cells. Mutations in Foxp3 cause severe, multi-organ autoimmunity in both human and mouse. FOXP3 can cooperate in a DNA-binding complex with NFAT (nuclear factor of activated T cells) to regulate the transcription of several known target genes. However, the global set of genes regulated directly by Foxp3 is not known and consequently, how this transcription factor controls the gene expression programme for T(reg) function is not understood. Here we identify Foxp3 target genes and report that many of these are key modulators of T-cell activation and function. Remarkably, the predominant, although not exclusive, effect of Foxp3 occupancy is to suppress the activation of target genes on T-cell stimulation. Foxp3 suppression of its targets appears to be crucial for the normal function of T(reg) cells, because overactive variants of some target genes are known to be associated with autoimmune disease.
Figures

References
-
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. - PubMed
-
- Baecher-Allan C, Hafler DA. Human regulatory T cells and their role in autoimmune disease. Immunol Rev. 2006;212:203–216. - PubMed
-
- Shevach EM. CD4+ CD25+ suppressor T cells: more questions than answers. Nature Rev Immunol. 2002;2:389–400. - PubMed
-
- von Boehmer H. Mechanisms of suppression by suppressor T cells. Nature Immunol. 2005;6:338–344. - PubMed
-
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

