Improved single-chain transactivators of the Tet-On gene expression system
- PMID: 17239234
- PMCID: PMC1797012
- DOI: 10.1186/1472-6750-7-6
Improved single-chain transactivators of the Tet-On gene expression system
Abstract
Background: The Tet-Off (tTA) and Tet-On (rtTA) regulatory systems are widely applied to control gene expression in eukaryotes. Both systems are based on the Tet repressor (TetR) from transposon Tn10, a dimeric DNA-binding protein that binds to specific operator sequences (tetO). To allow the independent regulation of multiple genes, novel Tet systems are being developed that respond to different effectors and bind to different tetO sites. To prevent heterodimerization when multiple Tet systems are expressed in the same cell, single-chain variants of the transactivators have been constructed. Unfortunately, the activity of the single-chain rtTA (sc-rtTA) is reduced when compared with the regular rtTA, which might limit its application.
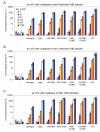
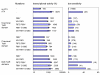
Results: We recently identified amino acid substitutions in rtTA that greatly improved the transcriptional activity and doxycycline-sensitivity of the protein. To test whether we can similarly improve other TetR-based gene regulation systems, we introduced these mutations into tTA and sc-rtTA. Whereas none of the tested mutations improved tTA activity, they did significantly enhance sc-rtTA activity. We thus generated a novel sc-rtTA variant that is almost as active and dox-sensitive as the regular dimeric rtTA. This variant was also less sensitive to interference by co-expressed TetR-based tTS repressor protein and may therefore be more suitable for applications where multiple TetR-based regulatory systems are used.
Conclusion: We developed an improved sc-rtTA variant that may replace regular rtTA in applications where multiple TetR-based regulatory systems are used.
Figures

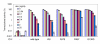

Similar articles
-
Generation and characterization of tTS-H4: a novel transcriptional repressor that is compatible with the reverse tetracycline-controlled TET-ON system.J Gene Med. 2007 Apr;9(4):308-18. doi: 10.1002/jgm.1012. J Gene Med. 2007. PMID: 17330923
-
Optimization of the Tet-On system for regulated gene expression through viral evolution.Gene Ther. 2006 Oct;13(19):1382-90. doi: 10.1038/sj.gt.3302780. Epub 2006 May 25. Gene Ther. 2006. PMID: 16724096
-
Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter.Nucleic Acids Res. 2005 Apr 4;33(6):e63. doi: 10.1093/nar/gni062. Nucleic Acids Res. 2005. PMID: 15809225 Free PMC article.
-
Tetracycline-controlled genetic switches.Handb Exp Pharmacol. 2007;(178):49-72. doi: 10.1007/978-3-540-35109-2_3. Handb Exp Pharmacol. 2007. PMID: 17203651 Review.
-
Tet-On Systems For Doxycycline-inducible Gene Expression.Curr Gene Ther. 2016;16(3):156-67. doi: 10.2174/1566523216666160524144041. Curr Gene Ther. 2016. PMID: 27216914 Free PMC article. Review.
Cited by
-
Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch.Nucleic Acids Res. 2008 Sep;36(16):e101. doi: 10.1093/nar/gkn443. Epub 2008 Jul 16. Nucleic Acids Res. 2008. PMID: 18632760 Free PMC article.
-
Establishment and Characterization of a Stable Producer Cell Line Generation Platform for the Manufacturing of Clinical-Grade Lentiviral Vectors.Biomedicines. 2024 Oct 4;12(10):2265. doi: 10.3390/biomedicines12102265. Biomedicines. 2024. PMID: 39457578 Free PMC article.
-
HIV-1 evolution: frustrating therapies, but disclosing molecular mechanisms.Philos Trans R Soc Lond B Biol Sci. 2010 Jun 27;365(1548):1965-73. doi: 10.1098/rstb.2010.0072. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20478891 Free PMC article. Review.
-
Multiscale effects of heating and cooling on genes and gene networks.Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):E10797-E10806. doi: 10.1073/pnas.1810858115. Epub 2018 Oct 19. Proc Natl Acad Sci U S A. 2018. PMID: 30341217 Free PMC article.
-
An improved Tet-on system in microRNA overexpression and CRISPR/Cas9-mediated gene editing.J Anim Sci Biotechnol. 2019 Jun 10;10:43. doi: 10.1186/s40104-019-0354-5. eCollection 2019. J Anim Sci Biotechnol. 2019. PMID: 31198556 Free PMC article.
References
-
- Gossen M, Bujard H. Tetracyclines in the control of gene expression in eukaryotes. In: Nelson M, Hillen W and Greenwald RA, editor. Tetracyclines in biology, chemistry and medicine. Basel, Birkhäuser Verlag; 2001. pp. 139–157.
-
- Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

