Mouse SNP Miner: an annotated database of mouse functional single nucleotide polymorphisms
- PMID: 17239255
- PMCID: PMC1797019
- DOI: 10.1186/1471-2164-8-24
Mouse SNP Miner: an annotated database of mouse functional single nucleotide polymorphisms
Abstract
Background: The mapping of quantitative trait loci in rat and mouse has been extremely successful in identifying chromosomal regions associated with human disease-related phenotypes. However, identifying the specific phenotype-causing DNA sequence variations within a quantitative trait locus has been much more difficult. The recent availability of genomic sequence from several mouse inbred strains (including C57BL/6J, 129X1/SvJ, 129S1/SvImJ, A/J, and DBA/2J) has made it possible to catalog DNA sequence differences within a quantitative trait locus derived from crosses between these strains. However, even for well-defined quantitative trait loci (<10 Mb) the identification of candidate functional DNA sequence changes remains challenging due to the high density of sequence variation between strains.
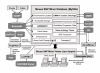
Description: To help identify functional DNA sequence variations within quantitative trait loci we have used the Ensembl annotated genome sequence to compile a database of mouse single nucleotide polymorphisms (SNPs) that are predicted to cause missense, nonsense, frameshift, or splice site mutations (available at http://bioinfo.embl.it/SnpApplet/). For missense mutations we have used the PolyPhen and PANTHER algorithms to predict whether amino acid changes are likely to disrupt protein function.
Conclusion: We have developed a database of mouse SNPs predicted to cause missense, nonsense, frameshift, and splice-site mutations. Our analysis revealed that 20% and 14% of missense SNPs are likely to be deleterious according to PolyPhen and PANTHER, respectively, and 6% are considered deleterious by both algorithms. The database also provides gene expression and functional annotations from the Symatlas, Gene Ontology, and OMIM databases to further assess candidate phenotype-causing mutations. To demonstrate its utility, we show that Mouse SNP Miner successfully finds a previously identified candidate SNP in the taste receptor, Tas1r3, that underlies sucrose preference in the C57BL/6J strain. We also use Mouse SNP Miner to derive a list of candidate phenotype-causing mutations within a previously uncharacterized QTL for response to morphine in the 129/Sv strain.
Figures

References
-
- Mouse Genome Informatics http://www.informatics.jax.org/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

