Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling
- PMID: 17239257
- PMCID: PMC1839132
- DOI: 10.1186/gb-2007-8-1-r12
Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling
Abstract
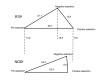
Background: T cells in the thymus undergo opposing positive and negative selection processes so that the only T cells entering circulation are those bearing a T cell receptor (TCR) with a low affinity for self. The mechanism differentiating negative from positive selection is poorly understood, despite the fact that inherited defects in negative selection underlie organ-specific autoimmune disease in AIRE-deficient people and the non-obese diabetic (NOD) mouse strain
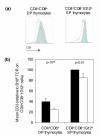
Results: Here we use homogeneous populations of T cells undergoing either positive or negative selection in vivo together with genome-wide transcription profiling on microarrays to identify the gene expression differences underlying negative selection to an Aire-dependent organ-specific antigen, including the upregulation of a genomic cluster in the cytogenetic band 2F. Analysis of defective negative selection in the autoimmune-prone NOD strain demonstrates a global impairment in the induction of the negative selection response gene set, but little difference in positive selection response genes. Combining expression differences with genetic linkage data, we identify differentially expressed candidate genes, including Bim, Bnip3, Smox, Pdrg1, Id1, Pdcd1, Ly6c, Pdia3, Trim30 and Trim12.
Conclusion: The data provide a molecular map of the negative selection response in vivo and, by analysis of deviations from this pathway in the autoimmune susceptible NOD strain, suggest that susceptibility arises from small expression differences in genes acting at multiple points in the pathway between the TCR and cell death.
Figures







Similar articles
-
Defective central tolerance induction in NOD mice: genomics and genetics.Immunity. 2005 Mar;22(3):385-96. doi: 10.1016/j.immuni.2005.01.015. Immunity. 2005. PMID: 15780994
-
Development of autoreactive diabetogenic T cells in the thymus of NOD mice.J Autoimmun. 2005 Feb;24(1):11-23. doi: 10.1016/j.jaut.2004.10.002. Epub 2004 Dec 8. J Autoimmun. 2005. PMID: 15725572
-
Hybridization signatures during thymus ontogeny reveals modulation of genes coding for T-cell signaling proteins.Mol Immunol. 2005 May;42(9):1043-8. doi: 10.1016/j.molimm.2004.09.031. Epub 2004 Nov 23. Mol Immunol. 2005. PMID: 15829294
-
Cross-talk between the T cell antigen receptor and the glucocorticoid receptor regulates thymocyte development.Stem Cells. 1996 Sep;14(5):490-500. doi: 10.1002/stem.140490. Stem Cells. 1996. PMID: 8888490 Review.
-
Genomic-scale analysis of gene expression in resting and activated T cells.Curr Opin Immunol. 2000 Apr;12(2):206-9. doi: 10.1016/s0952-7915(99)00075-8. Curr Opin Immunol. 2000. PMID: 10712937 Review.
Cited by
-
The transcriptional landscape of αβ T cell differentiation.Nat Immunol. 2013 Jun;14(6):619-32. doi: 10.1038/ni.2590. Epub 2013 May 5. Nat Immunol. 2013. PMID: 23644507 Free PMC article.
-
Selective ablation of thymic and peripheral Foxp3+ regulatory T cell development.Front Immunol. 2023 Dec 18;14:1298938. doi: 10.3389/fimmu.2023.1298938. eCollection 2023. Front Immunol. 2023. PMID: 38164128 Free PMC article.
-
Evasion of affinity-based selection in germinal centers by Epstein-Barr virus LMP2A.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11612-7. doi: 10.1073/pnas.1514484112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305967 Free PMC article.
-
Thymic negative selection is functional in NOD mice.J Exp Med. 2012 Mar 12;209(3):623-37. doi: 10.1084/jem.20112593. Epub 2012 Feb 13. J Exp Med. 2012. PMID: 22329992 Free PMC article.
-
Zeb1 represses TCR signaling, promotes the proliferation of T cell progenitors and is essential for NK1.1+ T cell development.Cell Mol Immunol. 2021 Sep;18(9):2140-2152. doi: 10.1038/s41423-020-0459-y. Epub 2020 May 12. Cell Mol Immunol. 2021. PMID: 32398809 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

