RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA
- PMID: 17239456
- PMCID: PMC1802126
- DOI: 10.1016/j.bbaexp.2006.11.006
RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA
Abstract
RNA interference (RNAi) is implicated in maintaining tandem DNA arrays as constitutive heterochromatin. We used chromatin immunoprecipitation with antibodies to RNA polymerase II (RNAPol-ChIP) to test for transcription of the following repeat arrays in human cells: subtelomeric D4Z4, pericentromeric satellite 2, and centromeric satellite alpha. D4Z4 has a promoter-like sequence upstream of an ORF in its 3.3-kb repeat unit. A short D4Z4 array at 4q35 is linked to facioscapulohumeral muscular dystrophy (FSHD). By RNAPol-ChIP and RT-PCR, little or no transcription of D4Z4 was detected in FSHD and normal myoblasts; lymphoblasts from an FSHD patient, a control, and a patient with D4Z4 hypomethylation due to mutation of DNMT3B (ICF syndrome); and normal or cancer tissues. However, RNAPol-ChIP assays indicated transcription of D4Z4 in a chromosome 4-containing human-mouse somatic cell hybrid. ChIP and RT-PCR showed satellite DNA transcription in some cancers and lymphoblastoid cell lines, although only at a low level. Given the evidence for the involvement of RNAi in satellite DNA heterochromatinization, it is surprising that, at most, a very small fraction of satellite DNA was associated with RNA Pol II. In addition, our results do not support the previously hypothesized disease-linked differential transcription of D4Z4 sequences in short, FSHD-linked arrays.
Figures


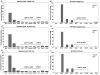


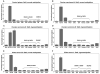
References
-
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. - PubMed
-
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki M, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. - PubMed
-
- Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome. 1995;6:76–83. - PubMed
-
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

