Application of microarrays to identify and characterize genes involved in attachment dependence in HeLa cells
- PMID: 17240181
- PMCID: PMC2001267
- DOI: 10.1016/j.ymben.2006.12.001
Application of microarrays to identify and characterize genes involved in attachment dependence in HeLa cells
Abstract
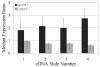
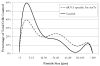
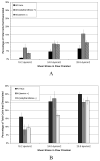
The ability to modify cellular properties such as adhesion is of interest in the design and performance of biotechnology-related processes. The current study was undertaken in order to evaluate the effectiveness of modulating cellular adhesion in HeLa cells from a genomics perspective. Using DNA microarrays, differences in gene expression between two phenotypically distinct, anchorage-dependent and anchorage-independent, HeLa cell lines were identified. With the aid of several statistical methods and an extensive literature search, two genes were selected as potential targets for further study: siat7e and lama4. Subsequently, experiments were carried out to investigate the effects of siat7e and lama4 separately, on adhesion in HeLa cells by altering their expression in vivo. Decreasing the expression of siat7e, a type II membrane glycosylating sialyltransferase, in anchorage-independent HeLa cells using short interfering RNA (siRNA) resulted in greater aggregation (i.e. clumping) and morphological changes as compared to untreated anchorage-independent HeLa cells. Similar effects were seen in anchorage-independent HeLa cells when the expression of lama4 which encodes laminin alpha4, a member of the laminin family of glycoproteins, was enhanced as compared to untreated anchorage-independent HeLa cells. Using a shear flow chamber, an attachment assay was developed; illustrating either increased expression of siat7e or decreased expression of lama4 in anchorage-dependent HeLa cells reduced cellular adhesion. Collectively, the results of this study are consistent with the roles siat7e and lama4 play in adhesion processes in vivo and indicate modifying the expression of either gene can influence adhesion in HeLa cells. The strategy of applying bioinformatics techniques to characterize and manipulate phenotypic behaviors is a powerful tool for altering the properties of various cell lines for desired biotechnology objectives.
Figures


Similar articles
-
Cells by design: a mini-review of targeting cell engineering using DNA microarrays.Mol Biotechnol. 2008 Jun;39(2):105-11. doi: 10.1007/s12033-008-9048-5. Mol Biotechnol. 2008. PMID: 18327555 Review.
-
Enhancement of cell proliferation in various mammalian cell lines by gene insertion of a cyclin-dependent kinase homolog.BMC Biotechnol. 2007 Oct 18;7:71. doi: 10.1186/1472-6750-7-71. BMC Biotechnol. 2007. PMID: 17945021 Free PMC article.
-
Stable Ectopic Expression of ST6GALNAC5 Induces Autocrine MET Activation and Anchorage-Independence in MDCK Cells.PLoS One. 2016 Feb 5;11(2):e0148075. doi: 10.1371/journal.pone.0148075. eCollection 2016. PLoS One. 2016. PMID: 26848584 Free PMC article.
-
Conversion of MDCK cell line to suspension culture by transfecting with human siat7e gene and its application for influenza virus production.Proc Natl Acad Sci U S A. 2009 Sep 1;106(35):14802-7. doi: 10.1073/pnas.0905912106. Epub 2009 Aug 17. Proc Natl Acad Sci U S A. 2009. PMID: 19706449 Free PMC article.
-
Gene expression under laser and light-emitting diodes radiation for modulation of cell adhesion: Possible applications for biotechnology.IUBMB Life. 2011 Sep;63(9):747-53. doi: 10.1002/iub.514. Epub 2011 Jul 27. IUBMB Life. 2011. PMID: 21796755 Review.
Cited by
-
Overexpression of ST6GalNAcV, a ganglioside-specific alpha2,6-sialyltransferase, inhibits glioma growth in vivo.Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12646-51. doi: 10.1073/pnas.0909862107. Epub 2010 Jun 28. Proc Natl Acad Sci U S A. 2010. PMID: 20616019 Free PMC article.
-
Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V.Molecules. 2015 Apr 16;20(4):6913-24. doi: 10.3390/molecules20046913. Molecules. 2015. PMID: 25913930 Free PMC article.
-
Cells by design: a mini-review of targeting cell engineering using DNA microarrays.Mol Biotechnol. 2008 Jun;39(2):105-11. doi: 10.1007/s12033-008-9048-5. Mol Biotechnol. 2008. PMID: 18327555 Review.
-
Viral vaccines and their manufacturing cell substrates: New trends and designs in modern vaccinology.Biotechnol J. 2015 Sep;10(9):1329-44. doi: 10.1002/biot.201400387. Epub 2015 Jul 24. Biotechnol J. 2015. PMID: 26212697 Free PMC article. Review.
-
Enhancement of cell proliferation in various mammalian cell lines by gene insertion of a cyclin-dependent kinase homolog.BMC Biotechnol. 2007 Oct 18;7:71. doi: 10.1186/1472-6750-7-71. BMC Biotechnol. 2007. PMID: 17945021 Free PMC article.
References
-
- Abulencia JP, Gaspard R, Healy ZR, Gaarde WA, Quackenbush J, Konstantopoulos K. Shear-induced cyclooxygenase-2 via a JNK2/c-Jun-dependent pathway regulates prostaglandin receptor expression in chondrocytic cells. J Biol Chem. 2003;278:28388–28394. - PubMed
-
- Bleckwenn NA, Bentley WE, Shiloach J. Evaluation of production parameters with the vaccinia virus expression system using microcarrier attached HeLa cells. Biotechnol Prog. 2005;21:554–561. - PubMed
-
- Brynda E, Pachernik J, Houska M, Pientka Z, Dvorak P. Surface immobilized protein multilayers for cell seeding. Langmuir. 2005;21:7877–7783. - PubMed
-
- Burke HB. Discovering Patterns in Microarray Data. Mol Diagn. 2000;5:349–357. - PubMed
-
- Butte A. The Use and Analysis of Microarray Data. Nat Rev Drug Discov. 2002;1:951–960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

