Chromosome-encoded narrow-spectrum Ambler class A beta-lactamase GIL-1 from Citrobacter gillenii
- PMID: 17242148
- PMCID: PMC1855525
- DOI: 10.1128/AAC.01152-06
Chromosome-encoded narrow-spectrum Ambler class A beta-lactamase GIL-1 from Citrobacter gillenii
Abstract
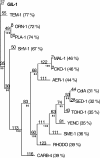
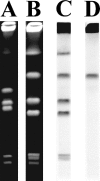
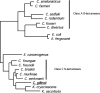
A novel beta-lactamase gene was cloned from the whole-cell DNA of an enterobacterial Citrobacter gillenii reference strain that displayed a weak narrow-spectrum beta-lactam-resistant phenotype and was expressed in Escherichia coli. It encoded a clavulanic acid-inhibited Ambler class A beta-lactamase, GIL-1, with a pI value of 7.5 and a molecular mass of ca. 29 kDa. GIL-1 had the highest percent amino acid sequence identity with TEM-1 and SHV-1, 77%, and 67%, respectively, and only 46%, 31%, and 32% amino acid sequence identity with CKO-1 (C. koseri), CdiA1 (C. diversus), and SED-1 (C. sedlaki), respectively. The substrate profile of the purified GIL-1 was similar to that of beta-lactamases TEM-1 and SHV-1. The blaGIL-1 gene was chromosomally located, as revealed by I-CeuI experiments, and was constitutively expressed at a low level in C. gillenii. No gene homologous to the regulatory ampR genes of chromosomal class C beta-lactamases was found upstream of the blaGIL-1 gene, which fits the noninducibility of beta-lactamase expression in C. gillenii. Rapid amplification of DNA 5' ends analysis of the promoter region revealed putative promoter sequences that diverge from what has been identified as the consensus sequence in E. coli. The blaGIL-1 gene was part of a 5.5-kb DNA fragment bracketed by a 9-bp duplication and inserted between the d-lactate dehydrogenase gene and the ydbH genes; this DNA fragment was absent in other Citrobacter species. This work further illustrates the heterogeneity of beta-lactamases in Citrobacter spp., which may indicate that the variability of Citrobacter species is greater than expected.
Figures


Similar articles
-
Inducible expression of the chromosomal cdiA from Citrobacter diversus NF85, encoding an ambler class A beta-lactamase, is under similar genetic control to the chromosomal ampC, encoding an ambler class C enzyme, from Citrobacter freundii OS60.Microb Drug Resist. 1995 Winter;1(4):285-91. doi: 10.1089/mdr.1995.1.285. Microb Drug Resist. 1995. PMID: 9158798
-
Characterization of the chromosomal class A beta-lactamase CKO from Citrobacter koseri.FEMS Microbiol Lett. 2006 Jan;254(2):285-92. doi: 10.1111/j.1574-6968.2005.00028.x. FEMS Microbiol Lett. 2006. PMID: 16445758
-
Citrobacter koseri and Citrobacter amalonaticus isolates carry highly divergent beta-lactamase genes despite having high levels of biochemical similarity and 16S rRNA sequence homology.J Antimicrob Chemother. 2004 Jun;53(6):1076-80. doi: 10.1093/jac/dkh235. Epub 2004 May 5. J Antimicrob Chemother. 2004. PMID: 15128725
-
Novel class A beta-lactamase Sed-1 from Citrobacter sedlakii: genetic diversity of beta-lactamases within the Citrobacter genus.Antimicrob Agents Chemother. 2001 Aug;45(8):2287-98. doi: 10.1128/AAC.45.8.2287-2298.2001. Antimicrob Agents Chemother. 2001. PMID: 11451687 Free PMC article.
-
[Mechanisms of endogenous drug resistance acquisition by spontaneous chromosomal gene mutation].Nihon Rinsho. 1997 May;55(5):1185-90. Nihon Rinsho. 1997. PMID: 9155173 Review. Japanese.
Cited by
-
Regulation of Class A β-Lactamase CzoA by CzoR and IscR in Comamonas testosteroni S44.Front Microbiol. 2017 Dec 22;8:2573. doi: 10.3389/fmicb.2017.02573. eCollection 2017. Front Microbiol. 2017. PMID: 29312251 Free PMC article.
-
Chromosomal integration and location on IncT plasmids of the blaCTX-M-2 gene in Proteus mirabilis clinical isolates.Antimicrob Agents Chemother. 2012 Feb;56(2):1093-6. doi: 10.1128/AAC.00258-11. Epub 2011 Nov 21. Antimicrob Agents Chemother. 2012. PMID: 22106217 Free PMC article.
-
Role of ISKpn7 and deletions in blaKPC gene expression.Antimicrob Agents Chemother. 2012 Sep;56(9):4753-9. doi: 10.1128/AAC.00334-12. Epub 2012 Jun 25. Antimicrob Agents Chemother. 2012. PMID: 22733068 Free PMC article.
-
Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate.Antimicrob Agents Chemother. 2010 Sep;54(9):3545-50. doi: 10.1128/AAC.00111-10. Epub 2010 Jun 21. Antimicrob Agents Chemother. 2010. PMID: 20566768 Free PMC article.
-
CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity.Antimicrob Agents Chemother. 2011 May;55(5):1861-6. doi: 10.1128/AAC.01656-10. Epub 2011 Feb 22. Antimicrob Agents Chemother. 2011. PMID: 21343457 Free PMC article.
References
-
- Avison, M. B., S. Underwood, A. Okazaki, T. R. Walsh, and P. M. Bennett. 2004. Analysis of AmpC beta-lactamase expression and sequence in biochemically atypical ceftazidime-resistant Enterobacteriaceae from paediatric patients. J. Antimicrob. Chemother. 53:584-591. - PubMed
-
- Brenner, D. J., C. M. O'Hara, P. A. Grimont, J. M. Janda, E. Falsen, E. Aldova, E. Ageron, J. Schindler, S. L. Abbott, and A. G. Steigerwalt. 1999. Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11). J. Clin. Microbiol. 37:2619-2624. - PMC - PubMed
-
- Brenner, D. J., P. A. D. Grimont, A. G. Stigerwalt, G. R. Fanning, E. Ageron, and C. F. Riddle. 1993. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmerii sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomespecies. Int. J. Syst. Bacteriol. 43:645-658. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Chemical Information
Molecular Biology Databases
Research Materials
Miscellaneous

