The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension
- PMID: 17242156
- PMCID: PMC1785119
- DOI: 10.1101/gad.1498707
The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension
Abstract
Sister chromatid cohesion, conferred by the evolutionarily conserved cohesin complex, is essential for proper chromosome segregation. Cohesin binds to discrete sites along chromosome arms, and is especially enriched surrounding centromeres, but past studies have not clearly defined the roles of arm and pericentromeric cohesion in chromosome segregation. To address this issue, we developed a technique that specifically reduced pericentromeric cohesin association on a single chromosome without affecting arm cohesin binding. Under these conditions, we observed more extensive stretching of centromeric chromatin and elevated frequencies of chromosome loss, suggesting that pericentromeric cohesin enrichment is essential for high-fidelity chromosome transmission. The magnitude of pericentromeric cohesin association was negatively correlated with tension between sister kinetochores, with the highest levels of association in cells lacking kinetochore-microtubule attachments. Pericentromeric cohesin recruitment required evolutionarily conserved components of the inner and central kinetochore. Together, these observations suggest that pericentromeric cohesin levels reflect the balance of opposing forces: the kinetochore-mediated enhancement of cohesin binding and the disruption of binding by mechanical tension at kinetochores. The involvement of conserved kinetochore components suggests that this pathway for pericentromeric cohesin enrichment may have been retained in higher eukaryotes to promote chromosome biorientation and accurate sister chromatid segregation.
Figures

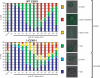

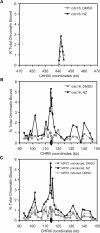

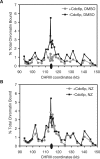
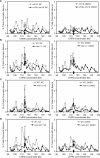
Comment in
-
Enhancing togetherness: kinetochores and cohesion.Genes Dev. 2007 Feb 1;21(3):238-41. doi: 10.1101/gad.1523107. Genes Dev. 2007. PMID: 17289914 Review. No abstract available.
References
-
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G., Ekwall K., Javerzat J.P., Cranston G., Javerzat J.P., Cranston G., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. - PubMed
-
- Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Partridge J.F., Genier S., Javerzat J.P., Allshire R.C., Genier S., Javerzat J.P., Allshire R.C., Javerzat J.P., Allshire R.C., Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. - PubMed
-
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Bhalla N., Sassoon I., Hyman A.A., Murray A.W., Sassoon I., Hyman A.A., Murray A.W., Hyman A.A., Murray A.W., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
