DNA methylation dictates histone H3K4 methylation
- PMID: 17242185
- PMCID: PMC1899905
- DOI: 10.1128/MCB.02291-06
DNA methylation dictates histone H3K4 methylation
Abstract
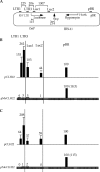
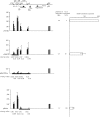
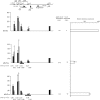
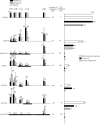
Histone lysine methylation and DNA methylation contribute to transcriptional regulation. We have previously shown that acetylated histones are associated with unmethylated DNA and are nearly absent from the methylated DNA regions by using patch-methylated stable episomes in human cells. The present study further demonstrates that DNA methylation immediately downstream from the transcription start site has a dramatic impact on transcription and that DNA methylation has a larger effect on transcription elongation than on initiation. We also show that dimethylated histone H3 at lysine 4 (H3K4me2) is depleted from regions with DNA methylation and that this effect is not linked to the transcriptional activity in the region. This effect is a local one and does not extend even 200 bp from the methylated DNA regions. Although depleted primarily from the methylated DNA regions, the presence of trimethylated histone H3 at lysine 4 (H3K4me3) may be affected by transcriptional activity as well. The data here suggest that DNA methylation at the junction of transcription initiation and elongation is most critical in transcription suppression and that this effect is mechanistically mediated through chromatin structure. The data also strongly support the model in which DNA methylation and not transcriptional activity dictates a closed chromatin structure, which excludes H3K4me2 and H3K4me3 in the region, as one of the pathways that safeguards the silent state of genes.
Figures
Similar articles
-
Transcriptional activity affects the H3K4me3 level and distribution in the coding region.Mol Cell Biol. 2010 Jun;30(12):2933-46. doi: 10.1128/MCB.01478-09. Epub 2010 Apr 19. Mol Cell Biol. 2010. PMID: 20404096 Free PMC article.
-
DNA methylation has a local effect on transcription and histone acetylation.Mol Cell Biol. 2002 Oct;22(19):6689-96. doi: 10.1128/MCB.22.19.6689-6696.2002. Mol Cell Biol. 2002. PMID: 12215526 Free PMC article.
-
Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells.Oncogene. 2008 Apr 10;27(17):2412-21. doi: 10.1038/sj.onc.1210895. Epub 2007 Oct 29. Oncogene. 2008. PMID: 17968314
-
Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation.Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194567. doi: 10.1016/j.bbagrm.2020.194567. Epub 2020 Apr 29. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32360393 Free PMC article. Review.
-
Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity?Epigenetics. 2009 Jul 1;4(5):302-6. doi: 10.4161/epi.4.5.9369. Epub 2009 Jul 25. Epigenetics. 2009. PMID: 19633430 Review.
Cited by
-
REPRODUCTIVE AGEING: Altered histone modification landscapes underpin defects in uterine stromal cell decidualization in aging females.Reproduction. 2024 Aug 2;168(3):e240171. doi: 10.1530/REP-24-0171. Print 2024 Sep 1. Reproduction. 2024. PMID: 38995736 Free PMC article.
-
Exposure to estrogen and ionizing radiation causes epigenetic dysregulation, activation of mitogen-activated protein kinase pathways, and genome instability in the mammary gland of ACI rats.Cancer Biol Ther. 2013 Jul;14(7):564-73. doi: 10.4161/cbt.24599. Epub 2013 May 10. Cancer Biol Ther. 2013. PMID: 23792640 Free PMC article.
-
Alterations of histone modifications at the senescence-associated gene HvS40 in barley during senescence.Plant Mol Biol. 2015 Sep;89(1-2):127-41. doi: 10.1007/s11103-015-0358-2. Epub 2015 Aug 7. Plant Mol Biol. 2015. PMID: 26249045
-
Quantitative Mass Spectrometry Reveals that Intact Histone H1 Phosphorylations are Variant Specific and Exhibit Single Molecule Hierarchical Dependence.Mol Cell Proteomics. 2016 Mar;15(3):818-33. doi: 10.1074/mcp.M114.046441. Epub 2015 Jul 24. Mol Cell Proteomics. 2016. PMID: 26209608 Free PMC article.
-
Epigenetic interplay between mouse endogenous retroviruses and host genes.Genome Biol. 2012 Oct 3;13(10):R89. doi: 10.1186/gb-2012-13-10-r89. Genome Biol. 2012. PMID: 23034137 Free PMC article.
References
-
- Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. - PubMed
-
- Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. - PubMed
-
- Cedar, H. 1988. DNA methylation and gene activity. Cell 53:3-4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
