Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases
- PMID: 17242192
- PMCID: PMC1899883
- DOI: 10.1128/MCB.01641-06
Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases
Abstract
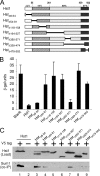
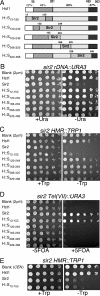
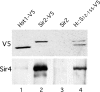
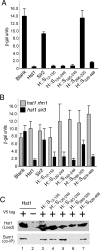
Sir2 and Hst1 are NAD(+)-dependent histone deacetylases of budding yeast that are related by strong sequence similarity. Nevertheless, the two proteins promote two mechanistically distinct forms of gene repression. Hst1 interacts with Rfm1 and Sum1 to repress the transcription of specific middle-sporulation genes. Sir2 interacts with Sir3 and Sir4 to silence genes contained within the silent-mating-type loci and telomere chromosomal regions. To identify the determinants of gene-specific versus regional repression, we created a series of Hst1::Sir2 hybrids. Our analysis yielded two dual-specificity chimeras that were able to perform both regional and gene-specific repression. Regional silencing by the chimeras required Sir3 and Sir4, whereas gene-specific repression required Rfm1 and Sum1. Our findings demonstrate that the nonconserved N-terminal region and two amino acids within the enzymatic core domain account for cofactor specificity and proper targeting of these proteins. These results suggest that the differences in the silencing and repression functions of Sir2 and Hst1 may not be due to differences in enzymatic activities of the proteins but rather may be the result of distinct cofactor specificities.
Figures


Similar articles
-
Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes.Mol Cell Biol. 2003 Mar;23(6):2009-16. doi: 10.1128/MCB.23.6.2009-2016.2003. Mol Cell Biol. 2003. PMID: 12612074 Free PMC article.
-
Genome-wide analysis of functional sirtuin chromatin targets in yeast.Genome Biol. 2013 May 27;14(5):R48. doi: 10.1186/gb-2013-14-5-r48. Genome Biol. 2013. PMID: 23710766 Free PMC article.
-
Sir Antagonist 1 (San1) is a ubiquitin ligase.J Biol Chem. 2004 Jun 25;279(26):26830-8. doi: 10.1074/jbc.M400894200. Epub 2004 Apr 12. J Biol Chem. 2004. PMID: 15078868
-
The Sir2 family of protein deacetylases.Annu Rev Biochem. 2004;73:417-35. doi: 10.1146/annurev.biochem.73.011303.073651. Annu Rev Biochem. 2004. PMID: 15189148 Review.
-
Identification and characterization of Sir2 inhibitors through phenotypic assays in yeast.Comb Chem High Throughput Screen. 2004 Nov;7(7):661-8. doi: 10.2174/1386207043328346. Comb Chem High Throughput Screen. 2004. PMID: 15578928 Review.
Cited by
-
Genealogy of an ancient protein family: the Sirtuins, a family of disordered members.BMC Evol Biol. 2013 Mar 5;13:60. doi: 10.1186/1471-2148-13-60. BMC Evol Biol. 2013. PMID: 23497088 Free PMC article.
-
Chromosome-wide histone deacetylation by sirtuins prevents hyperactivation of DNA damage-induced signaling upon replicative stress.Nucleic Acids Res. 2016 Apr 7;44(6):2706-26. doi: 10.1093/nar/gkv1537. Epub 2016 Jan 8. Nucleic Acids Res. 2016. PMID: 26748095 Free PMC article.
-
Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation.Mol Cells. 2009 Nov 30;28(5):407-15. doi: 10.1007/s10059-009-0169-x. Epub 2009 Nov 18. Mol Cells. 2009. PMID: 19936627 Free PMC article. Review.
-
Crystallization and preliminary crystallographic studies of the NAD+-dependent deacetylase HST1 from Saccharomyces cerevisiae.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Dec 1;67(Pt 12):1579-81. doi: 10.1107/S1744309111040589. Epub 2011 Nov 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 22139171 Free PMC article.
-
Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae.BMC Mol Biol. 2009 Jul 28;10:76. doi: 10.1186/1471-2199-10-76. BMC Mol Biol. 2009. PMID: 19638198 Free PMC article.
References
-
- Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan, and X. F. Zheng. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell. 10:1295-1305. - PubMed
-
- Avalos, J. L., I. Celic, S. Muhammad, M. S. Cosgrove, J. D. Boeke, and C. Wolberger. 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10:523-535. - PubMed
-
- Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. - PubMed
-
- Chang, J. H., H. C. Kim, K. Y. Hwang, J. W. Lee, S. P. Jackson, S. D. Bell, and Y. Cho. 2002. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 277:34489-34498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
