Human glucocorticoid receptor beta binds RU-486 and is transcriptionally active
- PMID: 17242213
- PMCID: PMC1820503
- DOI: 10.1128/MCB.01439-06
Human glucocorticoid receptor beta binds RU-486 and is transcriptionally active
Abstract
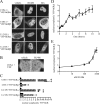
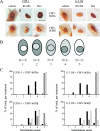
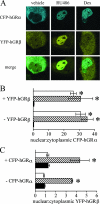
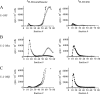
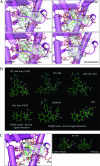
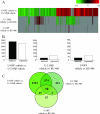
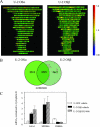
Human glucocorticoid receptor (hGR) is expressed as two alternately spliced C-terminal isoforms, alpha and beta. In contrast to the canonical hGRalpha, hGRbeta is a nucleus-localized orphan receptor thought not to bind ligand and not to affect gene transcription other than by acting as a dominant negative to hGRalpha. Here we used confocal microscopy to examine the cellular localization of transiently expressed fluorescent protein-tagged hGRbeta in COS-1 and U-2 OS cells. Surprisingly, yellow fluorescent protein (YFP)-hGRbeta was predominantly located in the cytoplasm and translocated to the nucleus following application of the glucocorticoid antagonist RU-486. This effect of RU-486 was confirmed with transiently expressed wild-type hGRbeta. Confocal microscopy of coexpressed YFP-hGRbeta and cyan fluorescent protein-hGRalpha in COS-1 cells indicated that the receptors move into the nucleus independently. Using a ligand binding assay, we confirmed that hGRbeta bound RU-486 but not the hGRalpha ligand dexamethasone. Examination of the cellular localization of YFP-hGRbeta in response to a series of 57 related compounds indicated that RU-486 is thus far the only identified ligand that interacts with hGRbeta. The selective interaction of RU-486 with hGRbeta was also supported by molecular modeling and computational docking studies. Interestingly, microarray analysis indicates that hGRbeta, expressed in the absence of hGRalpha, can regulate gene expression and furthermore that occupation of hGRbeta with the antagonist RU-486 diminishes that capacity despite the lack of helix 12 in the ligand binding domain.
Figures
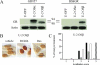
References
-
- Bledsoe, R. K., V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, and H. E. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. - PubMed
-
- Christodoulopoulos, P., D. Y. M. Leung, M. W. Elliott, J. C. Hogg, S. Muro, M. Toda, S. Laberge, and Q. Hamid. 2000. Increased number of glucocorticoid receptor-β-expressing cells in the airways in fatal asthma. J. Allergy Clin. Immunol. 106:479-484. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
