Evolution of mammalian CD1: marsupial CD1 is not orthologous to the eutherian isoforms and is a pseudogene in the opossum Monodelphis domestica
- PMID: 17244156
- PMCID: PMC2265927
- DOI: 10.1111/j.1365-2567.2007.02545.x
Evolution of mammalian CD1: marsupial CD1 is not orthologous to the eutherian isoforms and is a pseudogene in the opossum Monodelphis domestica
Abstract
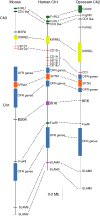
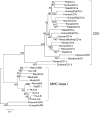
CD1 is a member of the major histocompatibility complex (MHC) class I family of proteins that present lipid antigens to T cells and natural killer (NK) T cells; it is found in both eutherian mammals and birds. In eutherians, duplication of the CD1 gene has resulted in multiple isoforms. A marsupial CD1 homologue was identified in a set of expressed sequence tags from the thymus of the bandicoot Isoodon macrourus. Southern blot and genomic sequence analyses revealed that CD1 is a single copy gene in both I. macrourus and a distantly related marsupial, the opossum Monodelphis domestica, which is currently the only marsupial species for which a whole genome sequence is available. We found that the opossum CD1 is located in a genomic region with a high degree of conserved synteny to the chromosomal regions containing human and mouse CD1. A phylogenetic analysis of mammalian CD1 revealed that marsupial CD1 is not orthologous to the eutherian CD1 isoforms, consistent with the latter having emerged by duplication after the separation of marsupials and eutherians 170-180 million years ago. The I. macrourus CD1 gene is actively transcribed and appears to encode a functional protein. In contrast, transcription of the M. domestica CD1 was not detected in any tissue and the predicted CD1 gene sequence contains a number of deletions that appear to render the locus a pseudogene.
Figures


References
-
- Jayawardena-Wolf J, Bendelac A. CD1 and lipid antigens: intracellular pathways for antigen presentation. Curr Opin Immunol. 2001;13:109–13. - PubMed
-
- Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–18. - PubMed
-
- Van Rhijn I, Koets AP, Im JS, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d1. J Immunol. 2006;176:4888–93. - PubMed
-
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

