RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha
- PMID: 17244529
- PMCID: PMC2563152
- DOI: 10.1016/j.molcel.2007.01.001
RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha
Abstract
Hypoxia-inducible factor 1 (HIF-1) regulates transcription in response to changes in O(2) concentration. O(2)-dependent degradation of the HIF-1alpha subunit is mediated by prolyl hydroxylase (PHD), the von Hippel-Lindau (VHL)/Elongin-C/Elongin-B E3 ubiquitin ligase complex, and the proteasome. Inhibition of heat-shock protein 90 (HSP90) leads to O(2)/PHD/VHL-independent degradation of HIF-1alpha. We have identified the receptor of activated protein kinase C (RACK1) as a HIF-1alpha-interacting protein that promotes PHD/VHL-independent proteasomal degradation of HIF-1alpha. RACK1 competes with HSP90 for binding to the PAS-A domain of HIF-1alpha in vitro and in human cells. HIF-1alpha degradation induced by the HSP90 inhibitor 17-allylaminogeldanamycin is abolished by RACK1 loss of function. RACK1 binds to Elongin-C and promotes ubiquitination of HIF-1alpha. Elongin-C-binding sites in RACK1 and VHL show significant sequence similarity. Thus, RACK1 is an essential component of an O(2)/PHD/VHL-independent mechanism for regulating HIF-1alpha stability through competition with HSP90 and recruitment of the Elongin-C/B ubiquitin ligase complex.
Figures


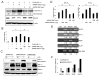



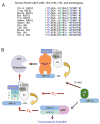
Similar articles
-
RACK1 vs. HSP90: competition for HIF-1 alpha degradation vs. stabilization.Cell Cycle. 2007 Mar 15;6(6):656-9. doi: 10.4161/cc.6.6.3981. Epub 2007 Mar 7. Cell Cycle. 2007. PMID: 17361105 Review.
-
Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization.J Biol Chem. 2007 Dec 21;282(51):37064-73. doi: 10.1074/jbc.M705015200. Epub 2007 Oct 26. J Biol Chem. 2007. PMID: 17965024 Free PMC article.
-
Spermidine/spermine N(1)-acetyltransferase-1 binds to hypoxia-inducible factor-1alpha (HIF-1alpha) and RACK1 and promotes ubiquitination and degradation of HIF-1alpha.J Biol Chem. 2007 Nov 16;282(46):33358-33366. doi: 10.1074/jbc.M705627200. Epub 2007 Sep 17. J Biol Chem. 2007. PMID: 17875644
-
COMMD1 Promotes pVHL and O2-Independent Proteolysis of HIF-1alpha via HSP90/70.PLoS One. 2009 Oct 5;4(10):e7332. doi: 10.1371/journal.pone.0007332. PLoS One. 2009. PMID: 19802386 Free PMC article.
-
Hypoxia-inducible factor 1 (HIF-1) pathway.Sci STKE. 2007 Oct 9;2007(407):cm8. doi: 10.1126/stke.4072007cm8. Sci STKE. 2007. PMID: 17925579 Review.
Cited by
-
The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions.Cell Div. 2016 May 23;11:7. doi: 10.1186/s13008-016-0020-7. eCollection 2016. Cell Div. 2016. PMID: 27222660 Free PMC article. Review.
-
STAT5 Is Necessary for the Metabolic Switch Induced by IL-2 in Cervical Cancer Cell Line SiHa.Int J Mol Sci. 2024 Jun 21;25(13):6835. doi: 10.3390/ijms25136835. Int J Mol Sci. 2024. PMID: 38999946 Free PMC article.
-
Hypoxia Inducible Factor 1 Alpha Is Expressed in Germ Cells throughout the Murine Life Cycle.PLoS One. 2016 May 5;11(5):e0154309. doi: 10.1371/journal.pone.0154309. eCollection 2016. PLoS One. 2016. PMID: 27148974 Free PMC article.
-
Heat Shock Protein 90 Associates with the Per-Arnt-Sim Domain of Heme-free Soluble Guanylate Cyclase: IMplications for Enzyme Maturation.J Biol Chem. 2015 Aug 28;290(35):21615-28. doi: 10.1074/jbc.M115.645515. Epub 2015 Jul 1. J Biol Chem. 2015. PMID: 26134567 Free PMC article.
-
Rack1 regulates B-cell development and function by binding to and stabilizing the transcription factor Pax5.Cell Mol Immunol. 2024 Nov;21(11):1282-1295. doi: 10.1038/s41423-024-01213-2. Epub 2024 Sep 10. Cell Mol Immunol. 2024. PMID: 39256480
References
-
- Baek JH, Mahon PC, Oh J, Kelly B, Krishnamachary B, Pearson M, Chan DA, Giaccia AJ, Semenza GL. OS-9 interacts with hypoxia-inducible factor 1α and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol Cell. 2005;17:503–512. - PubMed
-
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. - PubMed
-
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. - PubMed
-
- Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1α, HIF-2α, and other pathways. J Biol Chem. 2006;281:15215–15226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

