Gene network interconnectedness and the generalized topological overlap measure
- PMID: 17250769
- PMCID: PMC1797055
- DOI: 10.1186/1471-2105-8-22
Gene network interconnectedness and the generalized topological overlap measure
Abstract
Background: Network methods are increasingly used to represent the interactions of genes and/or proteins. Genes or proteins that are directly linked may have a similar biological function or may be part of the same biological pathway. Since the information on the connection (adjacency) between 2 nodes may be noisy or incomplete, it can be desirable to consider alternative measures of pairwise interconnectedness. Here we study a class of measures that are proportional to the number of neighbors that a pair of nodes share in common. For example, the topological overlap measure by Ravasz et al. 1 can be interpreted as a measure of agreement between the m = 1 step neighborhoods of 2 nodes. Several studies have shown that two proteins having a higher topological overlap are more likely to belong to the same functional class than proteins having a lower topological overlap. Here we address the question whether a measure of topological overlap based on higher-order neighborhoods could give rise to a more robust and sensitive measure of interconnectedness.
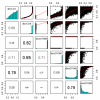
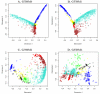
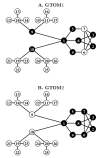
Results: We generalize the topological overlap measure from m = 1 step neighborhoods to m > or = 2 step neighborhoods. This allows us to define the m-th order generalized topological overlap measure (GTOM) by (i) counting the number of m-step neighbors that a pair of nodes share and (ii) normalizing it to take a value between 0 and 1. Using theoretical arguments, a yeast co-expression network application, and a fly protein network application, we illustrate the usefulness of the proposed measure for module detection and gene neighborhood analysis.
Conclusion: Topological overlap can serve as an important filter to counter the effects of spurious or missing connections between network nodes. The m-th order topological overlap measure allows one to trade-off sensitivity versus specificity when it comes to defining pairwise interconnectedness and network modules.
Figures




References
-
- Horvath S, Zhang B, Carlson M, Lu K, Zhu S, Felciano R, Laurance M, Zhao W, Shu Q, Lee Y, Scheck A, Liau L, Wu H, Geschwind D, Febbo P, Kornblum H, Cloughesy T, Nelson S, Mischel P. Analysis of Oncogenic Signaling Networks in Glioblastoma Identifies ASPM as a Novel Molecular Target. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

