Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication
- PMID: 17251286
- PMCID: PMC1866056
- DOI: 10.1128/JVI.02392-06
Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication
Abstract
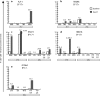
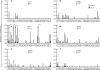
"Elite controllers" are individuals that durably control human immunodeficiency virus or simian immunodeficiency virus replication without therapeutic intervention. The study of these rare individuals may facilitate the definition of a successful immune response to immunodeficiency viruses. Here we describe six Indian-origin rhesus macaques that have controlled replication of the pathogenic virus SIVmac239 for 1 to 5 years. To determine which lymphocyte populations were responsible for this control, we transiently depleted the animals' CD8+ cells in vivo. This treatment resulted in 100- to 10,000-fold increases in viremia. When the CD8+ cells returned, control was reestablished and the levels of small subsets of previously subdominant CD8+ T cells expanded up to 2,500-fold above pre-depletion levels. This wave of CD8+ T cells was accompanied by robust Gag-specific CD4 responses. In contrast, CD8+ NK cell frequencies changed no more than threefold. Together, our data suggest that CD8+ T cells targeting a small number of epitopes, along with broad CD4+ T-cell responses, can successfully control the replication of the AIDS virus. It is likely that subdominant CD8+ T-cell populations play a key role in maintaining this control.
Figures




References
-
- Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O'Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361-4376. - PMC - PubMed
-
- Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069-7078. - PMC - PubMed
-
- Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738-749. - PMC - PubMed
-
- Barassi, C., A. Lazzarin, and L. Lopalco. 2004. CCR5-specific mucosal IgA in saliva and genital fluids of HIV-exposed seronegative subjects. Blood 104:2205-2206. - PubMed
-
- Betts, M. R., B. Exley, D. A. Price, A. Bansal, Z. T. Camacho, V. Teaberry, S. M. West, D. R. Ambrozak, G. Tomaras, M. Roederer, J. M. Kilby, J. Tartaglia, R. Belshe, F. Gao, D. C. Douek, K. J. Weinhold, R. A. Koup, P. Goepfert, and G. Ferrari. 2005. Characterization of functional and phenotypic changes in anti-Gag vaccine-induced T cell responses and their role in protection after HIV-1 infection. Proc. Natl. Acad. Sci. USA 102:4512-4517. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

