Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells
- PMID: 17251300
- PMCID: PMC1866039
- DOI: 10.1128/JVI.01552-06
Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells
Abstract
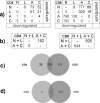
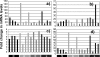
Changes in T-cell function are a hallmark of human immunodeficiency virus type 1 (HIV-1) infection, but the pathogenic mechanisms leading to these changes are unclear. We examined the gene expression profiles in ex vivo human CD4+ and CD8+ T cells from untreated HIV-1-infected individuals at different clinical stages and rates of disease progression. Profiles of pure CD4+ and CD8+ T-cell subsets from HIV-1-infected nonprogressors with controlled viremia were indistinguishable from those of individuals not infected with HIV-1. Similarly, no gene clusters could distinguish T cells from individuals with early infection from those seen in chronic progressive HIV-1 infection, whereas differences were observed between uninfected individuals or nonprogressors versus early or chronic progressors. In early and chronic HIV-1 infection, three characteristic gene expression signatures were observed. (i) CD4+ and CD8+ T cells showed increased expression of interferon-stimulated genes (ISGs). However, some ISGs, including CXCL9, CXCL10, and CXCL11, and the interleukin-15 alpha receptor were not upregulated. (ii) CD4+ and CD8+ T cells showed a cluster similar to that observed in thymocytes. (iii) More genes were differentially regulated in CD8+ T cells than in CD4+ T cells, including a cluster of genes downregulated exclusively in CD8+ T cells. In conclusion, HIV-1 infection induces a persistent T-cell transcriptional profile, early in infection, characterized by a dramatic but potentially aberrant interferon response and a profile suggesting an active thymic output. These findings highlight the complexity of the host-virus relationship in HIV-1 infection.
Figures




References
-
- Audigé, A., M. Urosevic, E. Schlaepfer, R. Walker, D. Powell, S. Hallenberger, H. Joller, H.-U. Simon, R. Dummer, and R. F. Speck. 2006. Anti-HIV state but not apoptosis depends on IFN signature in CD4+ T cells. J. Immunol. 177:6227-6237. - PubMed
-
- Bogdan, C., J. Mattner, and U. Schleicher. 2004. The role of type I interferons in non-viral infections. Immunol. Rev. 202:33-48. - PubMed
-
- Bosinger, S. E., K. A. Hosiawa, M. J. Cameron, D. Persad, L. Ran, L. Xu, M. R. Boulassel, M. Parenteau, J. Fournier, E. W. Rud, and D. J. Kelvin. 2004. Gene expression profiling of host response in models of acute HIV infection. J. Immunol. 173:6858-6863. - PubMed
-
- Brassard, D. L., M. M. Delorenzo, S. Cox, D. W. Leaman, Y. Sun, W. Ding, S. Gavor, J. Spond, F. Goodsaid, R. Bordens, and M. J. Grace. 2004. Regulation of gene expression by pegylated IFN-α2b and IFN-α2b in human peripheral blood mononuclear cells. J. Interferon Cytokine Res. 24:455-469. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

