Comparative estimation of the reproduction number for pandemic influenza from daily case notification data
- PMID: 17254982
- PMCID: PMC2358966
- DOI: 10.1098/rsif.2006.0161
Comparative estimation of the reproduction number for pandemic influenza from daily case notification data
Abstract
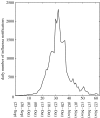
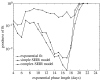
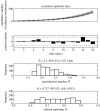
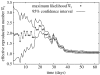
The reproduction number, R, defined as the average number of secondary cases generated by a primary case, is a crucial quantity for identifying the intensity of interventions required to control an epidemic. Current estimates of the reproduction number for seasonal influenza show wide variation and, in particular, uncertainty bounds for R for the pandemic strain from 1918 to 1919 have been obtained only in a few recent studies and are yet to be fully clarified. Here, we estimate R using daily case notifications during the autumn wave of the influenza pandemic (Spanish flu) in the city of San Francisco, California, from 1918 to 1919. In order to elucidate the effects from adopting different estimation approaches, four different methods are used: estimation of R using the early exponential-growth rate (Method 1), a simple susceptible-exposed-infectious-recovered (SEIR) model (Method 2), a more complex SEIR-type model that accounts for asymptomatic and hospitalized cases (Method 3), and a stochastic susceptible-infectious-removed (SIR) with Bayesian estimation (Method 4) that determines the effective reproduction number Rt at a given time t. The first three methods fit the initial exponential-growth phase of the epidemic, which was explicitly determined by the goodness-of-fit test. Moreover, Method 3 was also fitted to the whole epidemic curve. Whereas the values of R obtained using the first three methods based on the initial growth phase were estimated to be 2.98 (95% confidence interval (CI): 2.73, 3.25), 2.38 (2.16, 2.60) and 2.20 (1.55, 2.84), the third method with the entire epidemic curve yielded a value of 3.53 (3.45, 3.62). This larger value could be an overestimate since the goodness-of-fit to the initial exponential phase worsened when we fitted the model to the entire epidemic curve, and because the model is established as an autonomous system without time-varying assumptions. These estimates were shown to be robust to parameter uncertainties, but the theoretical exponential-growth approximation (Method 1) shows wide uncertainty. Method 4 provided a maximum-likelihood effective reproduction number 2.10 (1.21, 2.95) using the first 17 epidemic days, which is consistent with estimates obtained from the other methods and an estimate of 2.36 (2.07, 2.65) for the entire autumn wave. We conclude that the reproduction number for pandemic influenza (Spanish flu) at the city level can be robustly assessed to lie in the range of 2.0-3.0, in broad agreement with previous estimates using distinct data.
Figures



References
-
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans.
-
- Bettencourt, L. M. A. & Ribeiro, R. M. Submitted. Detecting early human transmission of H5N1 avian influenza. Proc. Natl Acad. Sci. USA.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

