Recombination and the nature of bacterial speciation
- PMID: 17255503
- PMCID: PMC2220085
- DOI: 10.1126/science.1127573
Recombination and the nature of bacterial speciation
Abstract
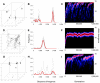
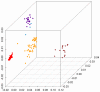
Genetic surveys reveal the diversity of bacteria and lead to the questioning of species concepts used to categorize bacteria. One difficulty in defining bacterial species arises from the high rates of recombination that results in the transfer of DNA between relatively distantly related bacteria. Barriers to this process, which could be used to define species naturally, are not apparent. Here, we review conceptual models of bacterial speciation and describe our computer simulations of speciation. Our findings suggest that the rate of recombination and its relation to genetic divergence have a strong influence on outcomes. We propose that a distinction be made between clonal divergence and sexual speciation. Hence, to make sense of bacterial diversity, we need data not only from genetic surveys but also from experimental determination of selection pressures and recombination rates and from theoretical models.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

