Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals
- PMID: 17257057
- PMCID: PMC1781497
- DOI: 10.1371/journal.pgen.0030019
Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals
Abstract
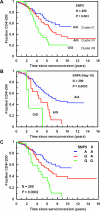
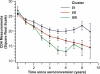
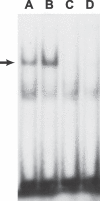
Human apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (Apobec3) antiretroviral factors cause hypermutation of proviral DNA leading to degradation or replication-incompetent HIV-1. However, HIV-1 viral infectivity factor (Vif) suppresses Apobec3 activity through the Cullin 5-Elongin B-Elongin C E3 ubiquitin ligase complex. We examined the effect of genetic polymorphisms in the CUL5 gene (encoding Cullin 5 protein) on AIDS disease progression in five HIV-1 longitudinal cohorts. A total of 12 single nucleotide polymorphisms (SNPs) spanning 93 kb in the CUL5 locus were genotyped and their haplotypes inferred. A phylogenetic network analysis revealed that CUL5 haplotypes were grouped into two clusters of evolutionarily related haplotypes. Cox survival analysis and mixed effects models were used to assess time to AIDS outcomes and CD4(+) T cell trajectories, respectively. Relative to cluster I haplotypes, the collective cluster II haplotypes were associated with more rapid CD4(+) T cell loss (relative hazards [RH] = 1.47 and p = 0.009), in a dose-dependent fashion. This effect was mainly attributable to a single cluster II haplotype (Hap10) (RH = 2.49 and p = 0.00001), possibly due to differential nuclear protein-binding efficiencies of a Hap10-specifying SNP as indicated by a gel shift assay. Consistent effects were observed for CD4(+) T cell counts and HIV-1 viral load trajectories over time. The findings of both functional and genetic epidemiologic consequences of CUL5 polymorphism on CD4(+) T cell and HIV-1 levels point to a role for Cullin 5 in HIV-1 pathogenesis and suggest interference with the Vif-Cullin 5 pathway as a possible anti-HIV-1 therapeutic strategy.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
-
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. - PubMed
-
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, et al. Species specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

