Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium
- PMID: 17257059
- PMCID: PMC1781477
- DOI: 10.1371/journal.ppat.0030007
Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium
Abstract
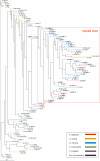
Enterococcus faecium, an ubiquous colonizer of humans and animals, has evolved in the last 15 years from an avirulent commensal to the third most frequently isolated nosocomial pathogen among intensive care unit patients in the United States. E. faecium combines multidrug resistance with the potential of horizontal resistance gene transfer to even more pathogenic bacteria. Little is known about the evolution and virulence of E. faecium, and genomic studies are hampered by the absence of a completely annotated genome sequence. To further unravel its evolution, we used a mixed whole-genome microarray and hybridized 97 E. faecium isolates from different backgrounds (hospital outbreaks (n = 18), documented infections (n = 34) and asymptomatic carriage of hospitalized patients (n = 15), and healthy persons (n = 15) and animals (n = 21)). Supported by Bayesian posterior probabilities (PP = 1.0), a specific clade containing all outbreak-associated strains and 63% of clinical isolates was identified. Sequencing of 146 of 437 clade-specific inserts revealed mobile elements (n = 74), including insertion sequence (IS) elements (n = 42), phage genes (n = 6) and plasmid sequences (n = 26), hypothetical (n = 58) and membrane proteins (n = 10), and antibiotic resistance (n = 9) and regulatory genes (n = 11), mainly located on two contigs of the unfinished E. faecium DO genome. Split decomposition analysis, varying guanine cytosine content, and aberrant codon adaptation indices all supported acquisition of these genes through horizontal gene transfer with IS16 as the predicted most prominent insert (98% sensitive, 100% specific). These findings suggest that acquisition of IS elements has facilitated niche adaptation of a distinct E. faecium subpopulation by increasing its genome plasticity. Increased genome plasticity was supported by higher diversity indices (ratio of average genetic similarities of pulsed-field gel electrophoresis and multi locus sequence typing) for clade-specific isolates. Interestingly, the previously described multi locus sequence typing-based clonal complex 17 largely overlapped with this clade. The present data imply that the global emergence of E. faecium, as observed since 1990, represents the evolution of a subspecies with a presumably better adaptation than other E. faecium isolates to the constraints of a hospital environment.
Conflict of interest statement
Figures



References
-
- National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–485. - PubMed
-
- Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. - PubMed
-
- Stobberingh E, van Den BA, London N, Driessen C, Top J, Willems R. Enterococci with glycopeptide resistance in turkeys, turkey farmers, turkey slaughterers, and (sub)urban residents in the south of The Netherlands: Evidence for transmission of vancomycin resistance from animals to humans? Antimicrob Agents Chemother. 1999;43:2215–2221. - PMC - PubMed
-
- van den Bogaard AE, Mertens P, London NH, Stobberingh EE. High prevalence of colonization with vancomycin- and pristinamycin-resistant enterococci in healthy humans and pigs in The Netherlands: Is the addition of antibiotics to animal feeds to blame? J Antimicrob Chemother. 1997;40:454–456. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

