Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency
- PMID: 17257062
- PMCID: PMC1781481
- DOI: 10.1371/journal.ppat.0030011
Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency
Abstract
A critical determinant in chronic gammaherpesvirus infections is the ability of these viruses to establish latency in a lymphocyte reservoir. The nuclear factor (NF)-kappaB family of transcription factors represent key players in B-cell biology and are targeted by gammaherpesviruses to promote host cell survival, proliferation, and transformation. However, the role of NF-kappaB signaling in the establishment of latency in vivo has not been addressed. Here we report the generation and in vivo characterization of a recombinant murine gammaherpesvirus 68 (gammaHV68) that expresses a constitutively active form of the NF-kappaB inhibitor, IkappaBalphaM. Inhibition of NF-kappaB signaling upon infection with gammaHV68-IkappaBalphaM did not affect lytic replication in cell culture or in the lung following intranasal inoculation. However, there was a substantial decrease in the frequency of latently infected lymphocytes in the lung (90% reduction) and spleens (97% reduction) 16 d post intranasal inoculation. Importantly, the defect in establishment of latency in lung B cells could not be overcome by increasing the dose of virus 100-fold. The observed decrease in establishment of viral latency correlated with a loss of activated, CD69(hi) B cells in both the lungs and spleen at day 16 postinfection, which was not apparent by 6 wk postinfection. Constitutive expression of Bcl-2 in B cells did not rescue the defect in the establishment of latency observed with gammaHV68-IkappaBalphaM, indicating that NF-kappaB-mediated functions apart from Bcl-2-mediated B-cell survival are critical for the efficient establishment of gammaherpesvirus latency in vivo. In contrast to the results obtained following intranasal inoculation, infection of mice with gammaHV68-IkappaBalphaM by the intraperitoneal route had only a modest impact on splenic latency, suggesting that route of inoculation may alter requirements for establishment of virus latency in B cells. Finally, analyses of the pathogenesis of gammaHV68-IkappaBalphaM provides evidence that NF-kappaB signaling plays an important role during multiple stages of gammaHV68 infection in vivo and, as such, represents a key host regulatory pathway that is likely manipulated by the virus to establish latency in B cells.
Conflict of interest statement
Figures
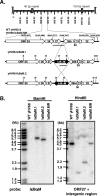
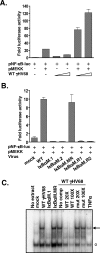

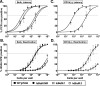




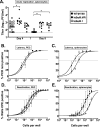
References
-
- Kim IJ, Flano E, Woodland DL, Lund FE, Randall TD, et al. Maintenance of long term gamma-herpesvirus B cell latency is dependent on CD40-mediated development of memory B cells. J Immunol. 2003;171:886–892. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 CA058524/CA/NCI NIH HHS/United States
- F32 AI066818/AI/NIAID NIH HHS/United States
- K12-GM000680/GM/NIGMS NIH HHS/United States
- R01 CA043143/CA/NCI NIH HHS/United States
- R01 CA052004/CA/NCI NIH HHS/United States
- AI58057/AI/NIAID NIH HHS/United States
- R01 AI058057/AI/NIAID NIH HHS/United States
- R01 CA95318/CA/NCI NIH HHS/United States
- K12 GM000680/GM/NIGMS NIH HHS/United States
- CA52004/CA/NCI NIH HHS/United States
- R01 CA095318/CA/NCI NIH HHS/United States
- CA58524/CA/NCI NIH HHS/United States
- R01 CA43143/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

