SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice
- PMID: 17259261
- PMCID: PMC1820975
- DOI: 10.1105/tpc.106.048124
SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice
Abstract
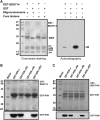
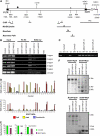
Although the role of H3K9 methylation in rice (Oryza sativa) is unclear, in Arabidopsis thaliana the loss of histone H3K9 methylation by mutation of Kryptonite [also known as SU(VAR)3-9 homolog] reduces genome-wide DNA methylation and increases the transcription of transposable elements. Here, we report that rice SDG714 (for SET Domain Group Protein714) encodes a histone H3K9-specific methyltransferase. The C terminus of SDG714 confers enzymatic activity and substrate specificity, whereas the N terminus localizes it in the nucleus. Loss-of-function mutants of SDG714 (SDG714IR transformants) generated by RNA interference display a mostly glabrous phenotype as a result of the lack of macro trichomes in glumes, leaves, and culms compared with control plants. These mutants also show decreased levels of CpG and CNG cytosine methylation as well as H3K9 methylation at the Tos17 locus, a copia-like retrotransposon widely used for the generation of rice mutants. Most interestingly, loss of function of SDG714 can enhance transcription and cause the transposition of Tos17. Together, these results suggest that histone H3K9 methylation mediated by SDG714 is involved in DNA methylation, the transposition of transposable elements, and genome stability in rice.
Figures





References
-
- Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13 627–637. - PubMed
-
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. - PubMed
-
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29 4319–4333. - PMC - PubMed
-
- Cao, X., Aufsatz, W., Zilberman, D., Mette, M.F., Huang, M.S., Matzke, M., and Jacobsen, S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13 2212–2217. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

