ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis
- PMID: 17259263
- PMCID: PMC1820965
- DOI: 10.1105/tpc.106.047761
ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis
Abstract
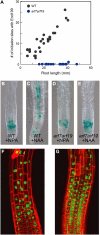
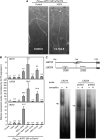
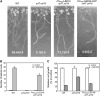
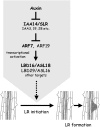
Lateral root formation in Arabidopsis thaliana is regulated by two related AUXIN RESPONSE FACTORs, ARF7 and ARF19, which are transcriptional activators of early auxin response genes. The arf7 arf19 double knockout mutant is severely impaired in lateral root formation. Target-gene analysis in arf7 arf19 transgenic plants harboring inducible forms of ARF7 and ARF19 revealed that ARF7 and ARF19 directly regulate the auxin-mediated transcription of LATERAL ORGAN BOUNDARIES-DOMAIN16/ASYMMETRIC LEAVES2-LIKE18 (LBD16/ASL18) and/or LBD29/ASL16 in roots. Overexpression of LBD16/ASL18 and LBD29/ASL16 induces lateral root formation in the absence of ARF7 and ARF19. These LBD/ASL proteins are localized in the nucleus, and dominant repression of LBD16/ASL18 activity inhibits lateral root formation and auxin-mediated gene expression, strongly suggesting that these LBD/ASLs function downstream of ARF7- and ARF19-dependent auxin signaling in lateral root formation. Our results reveal that ARFs regulate lateral root formation via direct activation of LBD/ASLs in Arabidopsis.
Figures


References
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251 533–549. - PubMed
-
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

