Deconvolving single-molecule intensity distributions for quantitative microscopy measurements
- PMID: 17259276
- PMCID: PMC1831712
- DOI: 10.1529/biophysj.106.101428
Deconvolving single-molecule intensity distributions for quantitative microscopy measurements
Abstract
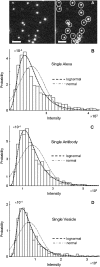
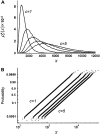
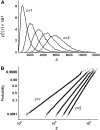
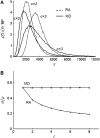
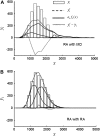
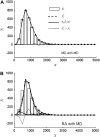
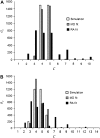
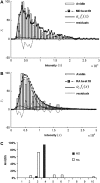
In fluorescence microscopy, images often contain puncta in which the fluorescent molecules are spatially clustered. This article describes a method that uses single-molecule intensity distributions to deconvolve the number of fluorophores present in fluorescent puncta as a way to "count" protein number. This method requires a determination of the correct statistical relationship between the single-molecule and single-puncta intensity distributions. Once the correct relationship has been determined, basis histograms can be generated from the single-molecule intensity distribution to fit the puncta distribution. Simulated data were used to demonstrate procedures to determine this relationship, and to test the methodology. This method has the advantages of single-molecule measurements, providing both the mean and variation in molecules per puncta. This methodology has been tested with the avidin-biocytin binding system for which the best-fit distribution of biocytins in the sample puncta was in good agreement with a bulk determination of the avidin-biocytin binding ratio.
Figures
References
-
- Selinummi, J., J. R. Sarkanen, A. Niemisto, M. L. Linne, T. Ylikomi, O. Yli-Harja, and T. O. Jalonen. 2006. Quantification of vesicles in differentiating human SH-SY5Y neuroblastoma cells by automated image analysis. Neurosci. Lett. 396:102–107. - PubMed
-
- Antonova, I., O. Arancio, A. C. Trillat, H. G. Wang, L. Zablow, H. Udo, E. R. Kandel, and R. D. Hawkins. 2001. Rapid increase in clusters of presynaptic proteins at onset of long-lasting potentiation. Science. 294:1547–1550. - PubMed
-
- Axelrod, D. 2001. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2:764–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

