Computational and analytical modeling of cationic lipid-DNA complexes
- PMID: 17259279
- PMCID: PMC1852371
- DOI: 10.1529/biophysj.106.096990
Computational and analytical modeling of cationic lipid-DNA complexes
Abstract
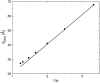
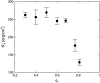
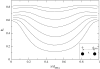
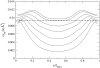
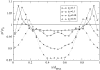
We present a theoretical study of the physical properties of cationic lipid-DNA (CL-DNA) complexes--a promising synthetically based nonviral carrier of DNA for gene therapy. The study is based on a coarse-grained molecular model, which is used in Monte Carlo simulations of mesoscopically large systems over timescales long enough to address experimental reality. In the present work, we focus on the statistical-mechanical behavior of lamellar complexes, which in Monte Carlo simulations self-assemble spontaneously from a disordered random initial state. We measure the DNA-interaxial spacing, d(DNA), and the local cationic area charge density, sigma(M), for a wide range of values of the parameter (c) representing the fraction of cationic lipids. For weakly charged complexes (low values of (c)), we find that d(DNA) has a linear dependence on (c)(-1), which is in excellent agreement with x-ray diffraction experimental data. We also observe, in qualitative agreement with previous Poisson-Boltzmann calculations of the system, large fluctuations in the local area charge density with a pronounced minimum of sigma(M) halfway between adjacent DNA molecules. For highly-charged complexes (large (c)), we find moderate charge density fluctuations and observe deviations from linear dependence of d(DNA) on (c)(-1). This last result, together with other findings such as the decrease in the effective stretching modulus of the complex and the increased rate at which pores are formed in the complex membranes, are indicative of the gradual loss of mechanical stability of the complex, which occurs when (c) becomes large. We suggest that this may be the origin of the recently observed enhanced transfection efficiency of lamellar CL-DNA complexes at high charge densities, because the completion of the transfection process requires the disassembly of the complex and the release of the DNA into the cytoplasm. Some of the structural properties of the system are also predicted by a continuum free energy minimization. The analysis, which semiquantitatively agrees with the computational results, shows that that mesoscale physical behavior of CL-DNA complexes is governed by interplay among electrostatic, elastic, and mixing free energies.
Figures



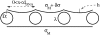
Similar articles
-
Transitions between distinct compaction regimes in complexes of multivalent cationic lipids and DNA.Biophys J. 2008 Jul;95(2):836-46. doi: 10.1529/biophysj.107.124669. Epub 2008 Apr 4. Biophys J. 2008. PMID: 18390608 Free PMC article.
-
Mesoscale computer modeling of lipid-DNA complexes for gene therapy.Phys Rev Lett. 2006 Jan 13;96(1):018102. doi: 10.1103/PhysRevLett.96.018102. Epub 2006 Jan 4. Phys Rev Lett. 2006. PMID: 16486522
-
Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes.Biophys J. 2003 May;84(5):3307-16. doi: 10.1016/S0006-3495(03)70055-1. Biophys J. 2003. PMID: 12719260 Free PMC article.
-
Cationic lipid-DNA complexes for gene therapy: understanding the relationship between complex structure and gene delivery pathways at the molecular level.Curr Med Chem. 2004 Jan;11(2):133-49. doi: 10.2174/0929867043456160. Curr Med Chem. 2004. PMID: 14754413 Review.
-
Modeling of cationic lipid-DNA complexes.Curr Med Chem. 2004 Jan;11(2):151-67. doi: 10.2174/0929867043456142. Curr Med Chem. 2004. PMID: 14754414 Review.
Cited by
-
Transitions between distinct compaction regimes in complexes of multivalent cationic lipids and DNA.Biophys J. 2008 Jul;95(2):836-46. doi: 10.1529/biophysj.107.124669. Epub 2008 Apr 4. Biophys J. 2008. PMID: 18390608 Free PMC article.
-
Relevance of the protein macrodipole in the membrane-binding process. Interactions of fatty-acid binding proteins with cationic lipid membranes.PLoS One. 2018 Mar 8;13(3):e0194154. doi: 10.1371/journal.pone.0194154. eCollection 2018. PLoS One. 2018. PMID: 29518146 Free PMC article.
-
Anomalous Diffusion of Polyelectrolyte Segments on Supported Charged Lipid Bilayers.Entropy (Basel). 2023 May 13;25(5):796. doi: 10.3390/e25050796. Entropy (Basel). 2023. PMID: 37238551 Free PMC article.
-
Elasticity and mechanical instability of charged lipid bilayers in ionic solutions.Eur Phys J E Soft Matter. 2014 Aug;37(8):26. doi: 10.1140/epje/i2014-14069-2. Epub 2014 Aug 15. Eur Phys J E Soft Matter. 2014. PMID: 25117501
-
Liquid crystalline phases of dendritic lipid-DNA self-assemblies: lamellar, hexagonal, and DNA bundles.J Phys Chem B. 2009 Mar 26;113(12):3694-703. doi: 10.1021/jp806863z. J Phys Chem B. 2009. PMID: 19673065 Free PMC article.
References
-
- Felgner, P. L., and G. Rhodes. 1991. Gene therapeutics. Nature. 349:351–352. - PubMed
-
- Smyth-Templeton, N., and D. D. Lasic, editors. 2000. Gene Therapy. Therapeutic Mechanisms and Strategies. Marcel Dekker, New York.
-
- Smith, A. E. 1995. Viral vectors in gene therapy. Annu. Rev. Microbiol. 49:807–838. - PubMed
-
- Kay, M. A., J. C. Glorioso, and L. Naldini. 2001. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 7:33–40. - PubMed
-
- Felgner, P. L., M. J. Heller, P. Lehn, J.-P. Behr, and F. C. Szoka, editors. 1996. Artificial self-assembling systems for gene delivery. American Chemical Society, Washington DC.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

