A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching
- PMID: 17259280
- PMCID: PMC1831683
- DOI: 10.1529/biophysj.106.096693
A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching
Abstract
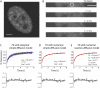
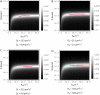
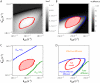
Fluorescence recovery after photobleaching (FRAP) is a powerful technique to study molecular dynamics inside living cells. During the past years, several laboratories have used FRAP to image the motion of RNA-protein and other macromolecular complexes in the nucleus and cytoplasm. In the case of mRNAs, there is growing evidence indicating that these molecules assemble into large ribonucleoprotein complexes that diffuse throughout the nucleus by Brownian motion. However, estimates of the corresponding diffusion rate yielded values that differ by up to one order of magnitude. In vivo labeling of RNA relies on indirect tagging with a fluorescent probe, and here we show how the binding affinity of the probe to the target RNA influences the effective diffusion estimates of the resulting complex. We extend current reaction-diffusion models for FRAP by allowing for diffusion of the bound complex. This more general model can be used to fit any fluorescence recovery curve involving two interacting mobile species in the cell (a fluorescent probe and its target substrate). The results show that interpreting FRAP data in light of the new model reconciles the discrepant mRNA diffusion-rate values previously reported.
Figures

References
-
- Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. - PubMed
-
- Lippincott-Schwartz, J., N. Altan-Bonnet, and G. H. Patterson. 2003. Photobleaching and photoactivation: following protein dynamics in living cells. Nat. Cell Biol. Suppl.:S7–S14. - PubMed
-
- Phair, R. D., and T. Misteli. 2001. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2:898–907. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

