The conformational landscape of the ribosomal protein S15 and its influence on the protein interaction with 16S RNA
- PMID: 17259282
- PMCID: PMC1831693
- DOI: 10.1529/biophysj.106.092601
The conformational landscape of the ribosomal protein S15 and its influence on the protein interaction with 16S RNA
Abstract
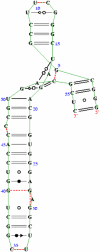
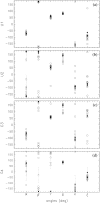
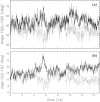
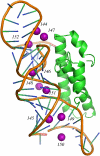
The interaction between the ribosomal protein S15 and its binding sites in the 16S RNA was examined from two points of view. First, the isolated protein S15 was studied by comparing NMR conformer sets, available in the PDB and recalculated using the CNS-ARIA protocol. Molecular dynamics (MD) trajectories were then recorded starting from a conformer of each set. The recalculation of the S15 NMR structure, as well as the recording of MD trajectories, reveals that several orientations of the N-terminal alpha-helix alpha1 with respect to the structure core are populated. MD trajectories of the complex between the ribosomal protein S15 and RNA were also recorded in the presence and absence of Mg(2+) ions. The Mg(2+) ions are hexacoordinated by water and RNA oxygens. The coordination spheres mainly interact with the RNA phosphodiester backbone, reducing the RNA mobility and inducing electrostatic screening. When the Mg(2+) ions are removed, the internal mobility of the RNA and of the protein increases at the interaction interface close to the RNA G-U/G-C motif as a result of a gap between the phosphate groups in the UUCG capping tetraloop and of the disruption of S15-RNA hydrogen bonds in that region. On the other hand, several S15-RNA hydrogen bonds are reinforced, and water bridges appear between the three-way junction region and S15. The network of hydrogen bonds observed in the loop between alpha1 and alpha2 is consequently reorganized. In the absence of Mg(2+), this network has the same pattern as the network observed in the isolated protein, where the helix alpha1 is mobile with respect to the protein core. The presence of Mg(2+) ions may thus play a role in stabilizing the orientation of the helix alpha1 of S15.
Figures






Similar articles
-
Binding interactions between the core central domain of 16S rRNA and the ribosomal protein S15 determined by molecular dynamics simulations.Nucleic Acids Res. 2003 Jan 15;31(2):629-38. doi: 10.1093/nar/gkg149. Nucleic Acids Res. 2003. PMID: 12527771 Free PMC article.
-
Molecular dynamics simulations of the denaturation and refolding of an RNA tetraloop.J Biomol Struct Dyn. 2001 Dec;19(3):381-96. doi: 10.1080/07391102.2001.10506748. J Biomol Struct Dyn. 2001. PMID: 11790138
-
Role of conserved nucleotides in building the 16 S rRNA binding site for ribosomal protein S15.J Mol Biol. 2001 Jan 26;305(4):785-803. doi: 10.1006/jmbi.2000.4354. J Mol Biol. 2001. PMID: 11162092
-
The 16S rRNA binding site of Thermus thermophilus ribosomal protein S15: comparison with Escherichia coli S15, minimum site and structure.RNA. 1996 Nov;2(11):1124-38. RNA. 1996. PMID: 8903343 Free PMC article.
-
A complex assembly landscape for the 30S ribosomal subunit.Annu Rev Biophys. 2009;38:197-215. doi: 10.1146/annurev.biophys.050708.133615. Annu Rev Biophys. 2009. PMID: 19416066 Free PMC article. Review.
Cited by
-
Role of salt-bridging interactions in recognition of viral RNA by arginine-rich peptides.Biophys J. 2021 Nov 16;120(22):5060-5073. doi: 10.1016/j.bpj.2021.10.007. Epub 2021 Oct 26. Biophys J. 2021. PMID: 34710377 Free PMC article.
-
Conformations of flanking bases in HIV-1 RNA DIS kissing complexes studied by molecular dynamics.Biophys J. 2007 Dec 1;93(11):3932-49. doi: 10.1529/biophysj.107.110056. Epub 2007 Aug 17. Biophys J. 2007. PMID: 17704156 Free PMC article.
-
RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview.Chem Rev. 2018 Apr 25;118(8):4177-4338. doi: 10.1021/acs.chemrev.7b00427. Epub 2018 Jan 3. Chem Rev. 2018. PMID: 29297679 Free PMC article. Review.
-
A computational investigation on the connection between dynamics properties of ribosomal proteins and ribosome assembly.PLoS Comput Biol. 2012;8(5):e1002530. doi: 10.1371/journal.pcbi.1002530. Epub 2012 May 24. PLoS Comput Biol. 2012. PMID: 22654657 Free PMC article.
-
Elastic properties of ribosomal RNA building blocks: molecular dynamics of the GTPase-associated center rRNA.Nucleic Acids Res. 2007;35(12):4007-17. doi: 10.1093/nar/gkm245. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553840 Free PMC article.
References
-
- Recht, M. I., and J. R. Williamson. 2004. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 344:395–407. - PubMed
-
- Nikulin, A., A. Serganov, E. Ennifar, S. Tishchenko, N. Nevskaya, W. Shepard, G. Portier, M. Garber, B. Ehresmann, C. Ehresmann, S. Nikonov, and P. Dumas. 2000. Crystal structure of the S15-rRNA complex. Nat. Struct. Biol. 7:273–277. - PubMed
-
- Ennifar, E., A. Nikulin, S. Tishchenko, A. Serganov, N. Nevskaya, M. Garber, B. Ehresmann, C. Ehresmann, S. Nikonov, and O. Dumas. 2000. The crystal structure of UUCG tetraloop. J. Mol. Biol. 304:34–42. - PubMed
-
- Berglund, H., A. Rak, A. Serganov, M. Garber, and T. Härd. 1997. Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus. Nat. Struct. Biol. 4:20–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

