A novel deaminase involved in chloronitrobenzene and nitrobenzene degradation with Comamonas sp. strain CNB-1
- PMID: 17259310
- PMCID: PMC1855817
- DOI: 10.1128/JB.01762-06
A novel deaminase involved in chloronitrobenzene and nitrobenzene degradation with Comamonas sp. strain CNB-1
Abstract
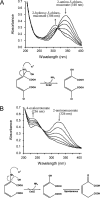
Comamonas sp. strain CNB-1 degrades nitrobenzene and chloronitrobenzene via the intermediates 2-aminomuconate and 2-amino-5-chloromuconate, respectively. Deamination of these two compounds results in the release of ammonia, which is used as a source of nitrogen for bacterial growth. In this study, a novel deaminase was purified from Comamonas strain CNB-1, and the gene (cnbZ) encoding this enzyme was cloned. The N-terminal sequence and peptide fingerprints of this deaminase were determined, and BLAST searches revealed no match with significant similarity to any functionally characterized proteins. The purified deaminase is a monomer (30 kDa), and its V(max) values for 2-aminomuconate and 2-amino-5-chloromuconate were 147 micromol x min(-1) x mg(-1) and 196 micromol x min(-1) x mg(-1), respectively. Its catalytic products from 2-aminomuconate and 2-amino-5-chloromuconate were 2-hydroxymuconate and 2-hydroxy-5-chloromuconate, respectively, which are different from those previously reported for the deaminases of Pseudomonas species. In the catalytic mechanism proposed, the alpha-carbon and nitrogen atoms (of both 2-aminomuconate and 2-amino-5-chloromuconate) were simultaneously attacked by a hydroxyl group and a proton, respectively. Homologs of cnbZ were identified in the genomes of Bradyrhizobium japonicum, Rhodopseudomonas palustris, and Roseiflexus sp. strain RS-1; these genes were previously annotated as encoding hypothetical proteins of unknown function. It is concluded that CnbZ represents a novel enzyme that deaminates xenobiotic compounds and/or alpha-amino acids.
Figures



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bhaumik, D., J. Medin, K. Gathy, and M. S. Coleman. 1993. Mutational analysis of active site residues of human adenosine deaminase. J. Biol. Chem. 268:5464-5470. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:249-254. - PubMed
-
- Denessiouk, K. A., A. I. Denesyuk, J. V. Lehtonen, T. Korpela, and M. S. Johnson. 1999. Common structural elements in the architecture of the cofactor-binding domains in unrelated families of pyridoxal phosphate-dependent enzymes. Proteins 35:250-261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

