Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses
- PMID: 17259356
- PMCID: PMC1828836
- DOI: 10.1128/AEM.01716-06
Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses
Abstract
The diversity and dynamics of bacterial populations in Saint-Nectaire, a raw-milk, semihard cheese, were investigated using a dual culture-dependent and direct molecular approach combining single-strand conformation polymorphism (SSCP) fingerprinting and sequencing of 16S rRNA genes. The dominant clones, among 125 16S rRNA genes isolated from milk, belonged to members of the Firmicutes (58% of the total clones) affiliated mainly with the orders Clostridiales and the Lactobacillales, followed by the phyla Proteobacteria (21.6%), Actinobacteria (16.8%), and Bacteroidetes (4%). Sequencing the 16S rRNA genes of 126 milk isolates collected from four culture media revealed the presence of 36 different species showing a wider diversity in the Gammaproteobacteria phylum and Staphylococcus genus than that found among clones. In cheese, a total of 21 species were obtained from 170 isolates, with dominant species belonging to the Lactobacillales and subdominant species affiliated with the Actinobacteria, Bacteroidetes (Chryseobacterium sp.), or Gammaproteobacteria (Stenotrophomonas sp.). Fingerprinting DNA isolated from milk by SSCP analysis yielded complex patterns, whereas analyzing DNA isolated from cheese resulted in patterns composed of a single peak which corresponded to that of lactic acid bacteria. SSCP fingerprinting of mixtures of all colonies harvested from plate count agar supplemented with crystal violet and vancomycin showed good potential for monitoring the subdominant Proteobacteria and Bacteroidetes (Flavobacteria) organisms in milk and cheese. Likewise, analyzing culturable subcommunities from cheese-ripening bacterial medium permitted assessment of the diversity of halotolerant Actinobacteria and Staphylococcus organisms. Direct and culture-dependent approaches produced complementary information, thus generating a more accurate view of milk and cheese microbial ecology.
Figures

 , F1 1-day cheese;
, F1 1-day cheese;  , F2 1-day cheese;
, F2 1-day cheese;  , F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.
, F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.
 , F1 1-day cheese;
, F1 1-day cheese;  , F2 1-day cheese;
, F2 1-day cheese;  , F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.
, F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.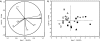
 , F1 1-day cheese;
, F1 1-day cheese;  , F2 1-day cheese;
, F2 1-day cheese;  , F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.
, F3 1-day cheese; ▴, F1 28-day cheese; •, F2 28-day cheese; ▪, F3 28-day cheese.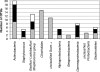
Similar articles
-
Stability of microbial communities in goat milk during a lactation year: molecular approaches.Syst Appl Microbiol. 2007 Nov;30(7):547-60. doi: 10.1016/j.syapm.2007.05.004. Epub 2007 Jul 2. Syst Appl Microbiol. 2007. PMID: 17604934
-
Comparative analysis of bacterial community composition in bulk tank raw milk by culture-dependent and culture-independent methods using the viability dye propidium monoazide.J Dairy Sci. 2014 Nov;97(11):6761-76. doi: 10.3168/jds.2014-8340. Epub 2014 Sep 18. J Dairy Sci. 2014. PMID: 25242425
-
Diversity and dynamics of lactobacilli populations during ripening of RDO Camembert cheese.Can J Microbiol. 2008 Mar;54(3):218-28. doi: 10.1139/w07-137. Can J Microbiol. 2008. PMID: 18388993
-
Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese.Int J Food Microbiol. 2011 Nov 1;150(2-3):81-94. doi: 10.1016/j.ijfoodmicro.2011.08.001. Epub 2011 Aug 8. Int J Food Microbiol. 2011. PMID: 21868118 Review.
-
Culture-independent methods for identifying microbial communities in cheese.Food Microbiol. 2008 Oct;25(7):839-48. doi: 10.1016/j.fm.2008.06.003. Epub 2008 Jun 10. Food Microbiol. 2008. PMID: 18721671 Review.
Cited by
-
Monitoring Changes in the Antimicrobial-Resistance Gene Set (ARG) of Raw Milk and Dairy Products in a Cattle Farm, from Production to Consumption.Vet Sci. 2024 Jun 8;11(6):265. doi: 10.3390/vetsci11060265. Vet Sci. 2024. PMID: 38922012 Free PMC article.
-
Adhesive properties of predominant bacteria in raw cow's milk to bovine mammary gland epithelial cells.Folia Microbiol (Praha). 2013 Nov;58(6):515-22. doi: 10.1007/s12223-013-0240-z. Epub 2013 Mar 27. Folia Microbiol (Praha). 2013. PMID: 23532507
-
Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage.Molecules. 2024 Jun 9;29(12):2745. doi: 10.3390/molecules29122745. Molecules. 2024. PMID: 38930811 Free PMC article.
-
Population dynamics of two antilisterial cheese surface consortia revealed by temporal temperature gradient gel electrophoresis.BMC Microbiol. 2010 Mar 11;10:74. doi: 10.1186/1471-2180-10-74. BMC Microbiol. 2010. PMID: 20222967 Free PMC article.
-
Assessment of bacterial and archaeal community structure in Swine wastewater treatment processes.Microb Ecol. 2015 Jul;70(1):77-87. doi: 10.1007/s00248-014-0537-8. Epub 2014 Nov 30. Microb Ecol. 2015. PMID: 25432577
References
-
- Andrighetto, C., E. Knijff, A. Lombardi, S. Torriani, M. Vancanneyt, K. Kersters, J. Swings, and F. Dellaglio. 2001. Phenotypic and genetic diversity of enterococci isolated from Italian cheeses. J. Dairy Res. 68:303-316. - PubMed
-
- Aquilanti, L., L. Dell'Aquila, E. Zannini, A. Zocchetti, and F. Clementi. 2006. Resident lactic acid bacteria in raw milk Canestrato Pugliese cheese. Lett. Appl. Microbiol. 43:161-167. - PubMed
-
- Beuvier, E., K. Berthaud, S. Cegarra, A. Dasen, S. Pochet, S. Buchin, and G. Duboz. 1997. Ripening and quality of swiss-type cheese made from raw, pasteurized or microfiltered milk. Int. Dairy J. 7:311-323.
-
- Brosius, J., T. J. Dull, D. Sleeter, and H. F. Noller. 1981. Gene organisation and primary structure of a ribosomal DNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

