DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins
- PMID: 17260987
- PMCID: PMC3677962
- DOI: 10.1021/ja065930i
DNA-encoded antibody libraries: a unified platform for multiplexed cell sorting and detection of genes and proteins
Abstract
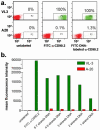
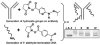
Whether for pathological examination or for fundamental biology studies, different classes of biomaterials and biomolecules are each measured from a different region of a typically heterogeneous tissue sample, thus introducing unavoidable sources of noise that are hard to quantitate. We describe the method of DNA-encoded antibody libraries (DEAL) for spatially multiplexed detection of ssDNAs and proteins as well as for cell sorting, all on the same diagnostic platform. DEAL is based upon the coupling of ssDNA oligomers onto antibodies which are then combined with the biological sample of interest. Spotted DNA arrays, which are found to inhibit biofouling, are utilized to spatially stratify the biomolecules or cells of interest. We demonstrate the DEAL technique for (1) the rapid detection of multiple proteins within a single microfluidic channel, and, with the additional step of electroless amplification of gold-nanoparticle labeled secondary antibodies, we establish a detection limit of 10 fM for the protein IL-2, 150 times more sensitive than the analogue ELISA; (2) the multiplexed, on-chip sorting of both immortalized cell lines and primary immune cells with an efficiency that exceeds surface-confined panning approaches; and (3) the co-detection of ssDNAs, proteins, and cell populations on the same platform.
Figures





References
-
- Lin B, White JT, Lu W, Xie T, Utleg AG, Yan X, Yi EC, Shannon P, Khretbukova I, Lange PH, Goodlett DR, Zhou D, Vasicek TJ, Hood L. Cancer Res. 2005;65:3081–3091. - PubMed
-
- Kwong KY, Bloom GC, Yang I, Boulware D, Coppola D, Haseman J, Chen E, McGrath A, Makusky AJ, Taylor J, Steiner S, Zhou J, Yeatman TJ, Quackenbush J. Genomics. 2005;86:142–158. - PubMed
-
- Huber M, Bahr I, Kratzchmar JR, Becker A, Muller E-C, Donner P, Pohlenz H-D, Schneider MR, Sommer A. Molec. Cell. Proteomics. 2004;3:43–55. - PubMed
-
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Molec. Cell. Proteomics. 2004;3:960–969. - PubMed
-
- Chen G, Gharib TG, Huang C-C, Taylor JMG, Misek DE, Kardia SLR, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Molec. Cell. Proteomics. 2002;1:304–313. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

