Electrochemiluminescent arrays for cytochrome P450-activated genotoxicity screening. DNA damage from benzo[a]pyrene metabolites
- PMID: 17261025
- PMCID: PMC2323608
- DOI: 10.1021/ac061975q
Electrochemiluminescent arrays for cytochrome P450-activated genotoxicity screening. DNA damage from benzo[a]pyrene metabolites
Erratum in
- Anal Chem. 2008 Mar 15;80(6):2272
Abstract
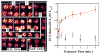
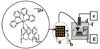
Arrays suitable for genotoxicity screening are reported that generate metabolites from cytochrome P450 enzymes (CYPs) in thin-film spots. Array spots containing DNA, various human cyt P450s, and electrochemiluminescence (ECL) generating metallopolymer [Ru(bpy)2PVP10]2+ were exposed to H2O2 to activate the enzymes. ECL from all spots was visualized simultaneously using a CCD camera. Using benzo[a]pyrene as a test substrate, enzyme activity for producing DNA damage in the arrays was found in the order CYP1B1 > CYP1A2 > CYP1A1 > CYP2E1 > myoglobin, the same as the order of their metabolic activity. Thus, these arrays estimate the relative propensity of different enzymes to produce genotoxic metabolites. This is the first demonstration of ECL arrays for high-throughput in vitro genotoxicity screening.
Figures
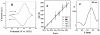






References
-
- Schenkman JB, Greim H, editors. Cytochrome P450. Springer-Verlag; Berlin: 1993.
-
- Ortiz de Montellano PR, editor. Cytochrome P450. Kluwer/Plenum; New York: 2005.
-
- Guengerich FP. Asia Pac J Pharmacol. 1990;5:327–345.
- Gonzalez FJ. Trends Pharm Sci. 1992;13:346–352. - PubMed
- Guengerich FP. Toxicol Lett. 1994;70:133–138. - PubMed
- Guengerich FP. Chem Res Toxicol. 2001;14:611–650. - PubMed
- Guengerich FP, Parikh A, Turesky RJ, Josephry PD. Mutat Res. 1999;428:115–124. - PubMed
- Guengerich FP. Chem Res Toxicol. 2001;14:611–650. - PubMed
-
- Tarun M, Rusling JF. Crit Rev Eukaryotic Gene Expression. 2005;15:295–315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

