Structural insights into fibronectin type III domain-mediated signaling
- PMID: 17261313
- PMCID: PMC2583400
- DOI: 10.1016/j.jmb.2006.10.017
Structural insights into fibronectin type III domain-mediated signaling
Abstract
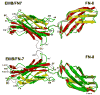
The alternatively spliced type III extradomain B (EIIIB) of fibronectin (FN) is expressed only during embryogenesis, wound healing and tumorigenesis. The biological function of this domain is unclear. We describe here the first crystal structure of the interface between alternatively spliced EIIIB and its adjacent FN type III domain 8 (FN B-8). The opened CC' loop of EIIIB, and the rotation and tilt of EIIIB allow good access to the FG loop of FN-8, which is normally hindered by the CC' loop of FN-7. In addition, the AGEGIP sequence of the CC'' loop of EIIIB replaces the NGQQGN sequence of the CC' loop of FN-7. Finally, the CC'' loop of EIIIB forms an acidic groove with FN-8. These structural findings warrant future studies directed at identifying potential binding partners for FN B-8 interface, linking EIIIB to skeletal and cartilaginous development, wound healing, and tumorigenesis, respectively.
Figures





Similar articles
-
Expression of alternatively spliced fibronectin variants during remodeling in proliferative glomerulonephritis.Am J Pathol. 1995 Nov;147(5):1361-71. Am J Pathol. 1995. PMID: 7485399 Free PMC article.
-
Plasma levels of fibronectin bearing the alternatively spliced EIIIB segment are increased after major trauma.J Lab Clin Med. 2003 Jun;141(6):401-10. doi: 10.1016/S0022-2143(03)00042-8. J Lab Clin Med. 2003. PMID: 12819638
-
Expression of the alternatively spliced EIIIB segment of fibronectin.Cell Adhes Commun. 1995 Feb;3(1):67-89. doi: 10.3109/15419069509081278. Cell Adhes Commun. 1995. PMID: 7538416
-
Alternative splicing of fibronectin: three variants, three functions.Bioessays. 1991 Oct;13(10):527-33. doi: 10.1002/bies.950131006. Bioessays. 1991. PMID: 1755828 Review.
-
[The alternative splicing & expression of fibronectin IIIcs segment and its relationship with wound healing].Fa Yi Xue Za Zhi. 2001 Nov;17(4):245-8. Fa Yi Xue Za Zhi. 2001. PMID: 12533877 Review. Chinese.
Cited by
-
Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein.J Biol Chem. 2012 May 18;287(21):17578-17588. doi: 10.1074/jbc.M111.303131. Epub 2012 Mar 22. J Biol Chem. 2012. PMID: 22442152 Free PMC article.
-
Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos.BMC Dev Biol. 2009 Jan 6;9:1. doi: 10.1186/1471-213X-9-1. BMC Dev Biol. 2009. PMID: 19126199 Free PMC article.
-
Nerve bundle formation during the promotion of peripheral nerve regeneration: collagen VI-neural cell adhesion molecule 1 interaction.Neural Regen Res. 2022 May;17(5):1023-1033. doi: 10.4103/1673-5374.324861. Neural Regen Res. 2022. PMID: 34558529 Free PMC article.
-
Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease.Hum Genet. 2021 Jan;140(1):77-111. doi: 10.1007/s00439-020-02147-1. Epub 2020 Mar 21. Hum Genet. 2021. PMID: 32200437 Review.
-
Structure and unfolding of the third type III domain from human fibronectin.Biochemistry. 2015 Nov 10;54(44):6724-33. doi: 10.1021/acs.biochem.5b00818. Epub 2015 Oct 30. Biochemistry. 2015. PMID: 26517579 Free PMC article.
References
-
- Hynes RO. Fibronectins. New York: Springer Verlag; 1990.
-
- George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development (Camb) 1993;119:1079–1091. - PubMed
-
- Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. - PubMed
-
- Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

