Biochemical characterization of an L-Xylulose reductase from Neurospora crassa
- PMID: 17261518
- PMCID: PMC1828828
- DOI: 10.1128/AEM.02515-06
Biochemical characterization of an L-Xylulose reductase from Neurospora crassa
Abstract
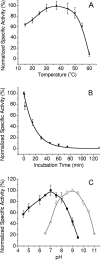
An l-xylulose reductase identified from the genome sequence of the filamentous fungus Neurospora crassa was heterologously expressed in Escherichia coli as a His(6) tag fusion protein, purified, and characterized. The enzyme may be used in the production of xylitol from the major pentose components of hemicellulosic waste, d-xylose and l-arabinose.
Figures

Similar articles
-
Characterization and gene cloning of l-xylulose reductase involved in l-arabinose catabolism from the pentose-fermenting fungus Rhizomucor pusillus.Biosci Biotechnol Biochem. 2017 Aug;81(8):1612-1618. doi: 10.1080/09168451.2017.1320518. Epub 2017 May 4. Biosci Biotechnol Biochem. 2017. PMID: 28471330
-
Heterologous expression, purification, and characterization of a highly active xylose reductase from Neurospora crassa.Appl Environ Microbiol. 2005 Mar;71(3):1642-7. doi: 10.1128/AEM.71.3.1642-1647.2005. Appl Environ Microbiol. 2005. PMID: 15746370 Free PMC article.
-
Cloning, characterization, and mutational analysis of a highly active and stable L-arabinitol 4-dehydrogenase from Neurospora crassa.Appl Microbiol Biotechnol. 2007 Dec;77(4):845-52. doi: 10.1007/s00253-007-1225-0. Epub 2007 Oct 16. Appl Microbiol Biotechnol. 2007. PMID: 17938906
-
Advances in applications, metabolism, and biotechnological production of L-xylulose.Appl Microbiol Biotechnol. 2016 Jan;100(2):535-40. doi: 10.1007/s00253-015-7087-y. Epub 2015 Nov 3. Appl Microbiol Biotechnol. 2016. PMID: 26526452 Review.
-
Polyol metabolism in fungi.Adv Microb Physiol. 1984;25:149-93. doi: 10.1016/s0065-2911(08)60292-1. Adv Microb Physiol. 1984. PMID: 6099966 Review. No abstract available.
Cited by
-
Data set of in silico simulation for the production of clavulanic acid and cephamycin C by Streptomyces clavuligerus using a genome scale metabolic model.Data Brief. 2019 May 15;24:103992. doi: 10.1016/j.dib.2019.103992. eCollection 2019 Jun. Data Brief. 2019. PMID: 31193725 Free PMC article.
-
Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa.Biotechnol Biofuels. 2017 Sep 21;10:225. doi: 10.1186/s13068-017-0901-2. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28947916 Free PMC article.
-
Identification of metabolic pathways expressed by Pichia anomala Kh6 in the presence of the pathogen Botrytis cinerea on apple: new possible targets for biocontrol improvement.PLoS One. 2014 Mar 10;9(3):e91434. doi: 10.1371/journal.pone.0091434. eCollection 2014. PLoS One. 2014. PMID: 24614090 Free PMC article.
-
Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides D-glucose, D-xylose and L-arabinose.Biotechnol Biofuels. 2014 Feb 28;7(1):31. doi: 10.1186/1754-6834-7-31. Biotechnol Biofuels. 2014. PMID: 24581151 Free PMC article.
References
-
- Chiang, C., and S. G. Knight. 1960. A new pathway of pentose metabolism. Biochem. Biophys. Res. Commun. 3:554-559. - PubMed
-
- El-Kabbani, O., V. Carbone, C. Darmanin, S. Ishikura, and A. Hara. 2005. Structure of the tetrameric form of human l-xylulose reductase: probing the inhibitor-binding site with molecular modeling and site-directed mutagenesis. Proteins 60:424-432. - PubMed
-
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. - PubMed
-
- Horer, S., J. Stoop, H. Mooibroek, U. Baumann, and J. Sassoon. 2001. The crystallographic structure of the mannitol 2-dehydrogenase NADP+ binary complex from Agaricus bisporus. J. Biol. Chem. 276:27555-27561. - PubMed
-
- Hutcheson, R. M., V. H. Reynolds, and O. Touster. 1956. The reduction of l-xylulose to xylitol by guinea pig liver mitochondria. J. Biol. Chem. 221:697-709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

