The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif
- PMID: 17261809
- PMCID: PMC1794263
- DOI: 10.1073/pnas.0607216104
The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif
Abstract
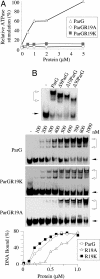
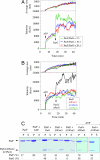
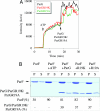
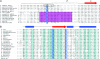
The ParF protein of plasmid TP228 belongs to the ubiquitous superfamily of ParA ATPases that drive DNA segregation in bacteria. ATP-bound ParF polymerizes into multistranded filaments. The partner protein ParG is dimeric, consisting of C-termini that interweave into a ribbon-helix-helix domain contacting the centromeric DNA and unstructured N-termini. ParG stimulates ATP hydrolysis by ParF approximately 30-fold. Here, we establish that the mobile tails of ParG are crucial for this enhancement and that arginine R19 within the tail is absolutely required for activation of ParF nucleotide hydrolysis. R19 is part of an arginine finger-like loop in ParG that is predicted to intercalate into the ParF nucleotide-binding pocket thereby promoting ATP hydrolysis. Significantly, mutations of R19 abrogated DNA segregation in vivo, proving that intracellular stimulation of ATP hydrolysis by ParG is a key regulatory process for partitioning. Furthermore, ParG bundles ParF-ATP filaments as well as promoting nucleotide-independent polymerization. The N-terminal flexible tail is required for both activities, because N-terminal DeltaParG polypeptides are defective in both functions. Strikingly, the critical arginine finger-like residue R19 is dispensable for ParG-mediated remodeling of ParF polymers, revealing that the ParG N-terminal tail possesses two separable activities in the interplay with ParF: a catalytic function during ATP hydrolysis and a mechanical role in modulation of polymerization. We speculate that activation of nucleotide hydrolysis via an arginine finger loop may be a conserved, regulatory mechanism of ParA family members and their partner proteins, including ParA-ParB and Soj-Spo0J that mediate DNA segregation and MinD-MinE that determine septum localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

