The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target
- PMID: 17263143
- PMCID: PMC3571657
The hepatitis C virus NS3 protein: a model RNA helicase and potential drug target
Abstract
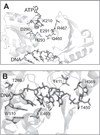
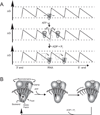
The C-terminal portion of hepatitis C virus (HCV) nonstructural protein 3 (NS3) forms a three domain polypeptide that possesses the ability to travel along RNA or single-stranded DNA (ssDNA) in a 3' to 5' direction. Fueled byATP hydrolysis, this movement allows the protein to displace complementary strands of DNA or RNA and proteins bound to the nucleic acid. HCV helicase shares two domains common to other motor proteins, one of which appears to rotate upon ATP binding. Several models have been proposed to explain how this conformational change leads to protein movement and RNA unwinding, but no model presently explains all existing experimental data. Compounds recently reported to inhibit HCV helicase, which include numerous small molecules, RNA aptamers and antibodies, will be useful for elucidating the role of a helicase in positive-sense single-stranded RNA virus replication and might serve as templates for the design of novel antiviral drugs.
Figures


Similar articles
-
HCV Helicase: Structure, Function, and Inhibition.In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 7. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience; 2006. Chapter 7. PMID: 21250378 Free Books & Documents. Review.
-
Structure and function of hepatitis C virus NS3 helicase.Curr Top Microbiol Immunol. 2000;242:171-96. doi: 10.1007/978-3-642-59605-6_9. Curr Top Microbiol Immunol. 2000. PMID: 10592661 Review.
-
In vitro selection of RNA aptamers against the HCV NS3 helicase domain.Oligonucleotides. 2004;14(2):114-29. doi: 10.1089/1545457041526335. Oligonucleotides. 2004. PMID: 15294075
-
Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor.Biochemistry. 2010 Mar 9;49(9):1822-32. doi: 10.1021/bi901974a. Biochemistry. 2010. PMID: 20108979 Free PMC article.
-
Docking studies of Pakistani HCV NS3 helicase: a possible antiviral drug target.PLoS One. 2014 Sep 4;9(9):e106339. doi: 10.1371/journal.pone.0106339. eCollection 2014. PLoS One. 2014. PMID: 25188400 Free PMC article.
Cited by
-
Benzothiazole and Pyrrolone Flavivirus Inhibitors Targeting the Viral Helicase.ACS Infect Dis. 2015 Mar 13;1(3):140-148. doi: 10.1021/id5000458. ACS Infect Dis. 2015. PMID: 26029739 Free PMC article.
-
Simultaneously Targeting the NS3 Protease and Helicase Activities for More Effective Hepatitis C Virus Therapy.ACS Chem Biol. 2015 Aug 21;10(8):1887-96. doi: 10.1021/acschembio.5b00101. Epub 2015 May 22. ACS Chem Biol. 2015. PMID: 25961497 Free PMC article.
-
Role of the Conserved DECH-Box Cysteine in Coupling Hepatitis C Virus Helicase-Catalyzed ATP Hydrolysis to RNA Unwinding.Biochemistry. 2018 Oct 30;57(43):6247-6255. doi: 10.1021/acs.biochem.8b00796. Epub 2018 Oct 16. Biochemistry. 2018. PMID: 30281972 Free PMC article.
-
Identification and biochemical characterization of halisulfate 3 and suvanine as novel inhibitors of hepatitis C virus NS3 helicase from a marine sponge.Mar Drugs. 2014 Jan 21;12(1):462-76. doi: 10.3390/md12010462. Mar Drugs. 2014. PMID: 24451189 Free PMC article.
-
Computational biology approach to uncover hepatitis C virus helicase operation.World J Gastroenterol. 2014 Apr 7;20(13):3401-9. doi: 10.3748/wjg.v20.i13.3401. World J Gastroenterol. 2014. PMID: 24707123 Free PMC article. Review.
References
-
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 1997;4:686–689. - PubMed
-
- Artsaenko O, Tessmann K, Sack M, Haussinger D, Heintges T. Abrogation of hepatitis C virus NS3 helicase enzymatic activity by recombinant human antibodies. J. Gen. Virol. 2003;84:2323–2332. - PubMed
-
- Astumian RD. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

