Relative information content and top-down proteomics by mass spectrometry: utility of ion/ion proton-transfer reactions in electrospray-based approaches
- PMID: 17263338
- PMCID: PMC2575742
- DOI: 10.1021/ac061798t
Relative information content and top-down proteomics by mass spectrometry: utility of ion/ion proton-transfer reactions in electrospray-based approaches
Abstract
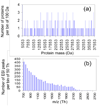
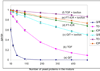
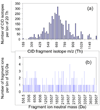
Computer simulations of electrospray ionization (ESI) and collision-induced dissociation (CID) experiments were employed to examine the informing power associated with "top-down" proteomics implemented with some commonly used mass analyzers, i.e., the quadrupole ion trap (QIT), the Fourier transform-ion cyclotron resonance mass spectrometer (FT-ICRMS), and the time-of-flight (TOF) mass spectrometer. Using a ratio of the separated (or resolved) peaks to the total number of predicted peaks as a measure of informing power, the ESI-MS simulation of a mixture of proteins showed that the FT-ICRMS exhibited the highest informing power among the three instruments being studied, with the QIT giving the lowest informing power, which was expected from the analysis of the "component capacity" of the three approaches. Also as expected on the basis of resolving elements per component, a dramatic increase in the informing power of the approach was obtained when ion/ion proton-transfer reactions were used to reduce the number of peaks and to minimize overlap between ions of different mass and charge but similar mass-to-charge ratio. With the assumptions made in this study, the informing power of the TOF + ion/ion approach rivaled or even exceeded that of the FT-ICRMS approach, despite significantly lower mass resolution. This result stemmed from both a reduction in the number of peaks and their dispersion over a much wider range of mass-to-charge ratios. Similar results were obtained from the CID simulation, where the informing power of different approaches was evaluated on the basis of the ratio of the number of ions for which a mass could be determined unambiguously to the total number of ions in the spectra.
Figures




References
-
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. - PubMed
-
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Mass Spectrom. Rev. 1990;9:37–70.
-
- Smith RD, Loo JA, Edmonds CG, Barinaga CJ, Udseth HR. Anal. Chem. 1990;62:882–899. - PubMed
-
- Busman M, Rockwood AL, Smith RD. J. Phys. Chem. 1992;96:2397–2400.
-
- Rockwood AL, Busman M, Smith RD. Int. J. Mass Spectrom. Ion Processes. 1991;111:103–129.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

