The beta-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6
- PMID: 17263687
- PMCID: PMC1876377
- DOI: 10.1042/BJ20070053
The beta-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6
Abstract
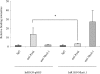
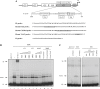
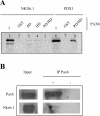
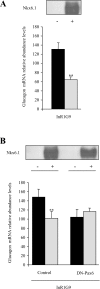
The transcription factor Nkx6.1 is required for the establishment of functional insulin-producing beta-cells in the endocrine pancreas. Overexpression of Nkx6.1 has been shown to inhibit glucagon gene expression while favouring insulin gene activation. Down-regulation resulted in the opposite effect, suggesting that absence of Nkx6.1 favours glucagon gene expression. To understand the mechanism by which Nkx6.1 suppresses glucagon gene expression, we studied its effect on the glucagon gene promoter activity in non-islet cells using transient transfections and gel-shift analyses. In glucagonoma cells transfected with an Nkx6.1-encoding vector, the glucagon promoter activity was reduced by 65%. In BHK21 cells, Nkx6.1 inhibited by 93% Pax6-mediated activation of the glucagon promoter, whereas Cdx2/3 and Maf stimulations were unaltered. Although Nkx6.1 could interact with both the G1 and G3 element, only the former displayed specificity for Nkx6.1. Mutagenesis of the three potential AT-rich motifs within the G1 revealed that only the Pax6-binding site preferentially interacted with Nkx6.1. Chromatin immunoprecipitation confirmed interaction of Nkx6.1 with the glucagon promoter and revealed a direct competition for binding between Pax6 and Nkx6.1. A weak physical interaction between Pax6 and Nkx6.1 was detected in vitro and in vivo suggesting that Nkx6.1 predominantly inhibits glucagon gene transcription through G1-binding competition. We suggest that cell-specific expression of the glucagon gene may only proceed when Nkx6.1, in combination with Pdx1 and Pax4, are silenced in early alpha-cell precursors.
Figures


References
-
- Habener J. F., Kemp D. M., Thomas M. K. Minireview: transcriptional regulation in pancreatic development. Endocrinology. 2005;146:1025–1034. - PubMed
-
- Johnson J., Carisson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. - PubMed
-
- Li H., Arber S., Jessel T. M., Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat. Genet. 1999;23:67–70. - PubMed
-
- Harrison K. A., Thaler J., Pfaff S. L., Gu H., Kehrl J. H. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat. Genet. 1999;23:71–75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

