DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein
- PMID: 17264121
- PMCID: PMC1851648
- DOI: 10.1093/nar/gkm001
DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein
Abstract
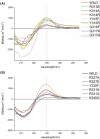
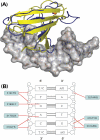
WRKY proteins, defined by the conserved WRKYGQK sequence, are comprised of a large superfamily of transcription factors identified specifically from the plant kingdom. This superfamily plays important roles in plant disease resistance, abiotic stress, senescence as well as in some developmental processes. In this study, the Arabidopsis WRKY1 was shown to be involved in the salicylic acid signaling pathway and partially dependent on NPR1; a C-terminal domain of WRKY1, AtWRKY1-C, was constructed for structural studies. Previous investigations showed that DNA binding of the WRKY proteins was localized at the WRKY domains and these domains may define novel zinc-binding motifs. The crystal structure of the AtWRKY1-C determined at 1.6 A resolution has revealed that this domain is composed of a globular structure with five beta strands, forming an antiparallel beta-sheet. A novel zinc-binding site is situated at one end of the beta-sheet, between strands beta4 and beta5. Based on this high-resolution crystal structure and site-directed mutagenesis, we have defined and confirmed that the DNA-binding residues of AtWRKY1-C are located at beta2 and beta3 strands. These results provided us with structural information to understand the mechanism of transcriptional control and signal transduction events of the WRKY proteins.
Figures

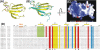

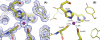
References
-
- Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. - PubMed
-
- Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol. Genet. Genomics. 2002;267:154–161. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

