Inhibition of MDR1 expression with altritol-modified siRNAs
- PMID: 17264131
- PMCID: PMC1851637
- DOI: 10.1093/nar/gkl1126
Inhibition of MDR1 expression with altritol-modified siRNAs
Abstract
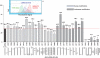
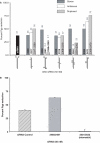
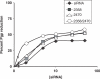
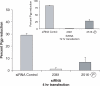
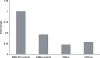
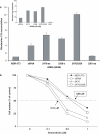
Altritol-modified nucleic acids (ANAs) support RNA-like A-form structures when included in oligonucleotide duplexes. Thus altritol residues seem suitable as candidates for the chemical modification of siRNAs. Here we report that ANA-modified siRNAs targeting the MDR1 gene can exhibit improved efficacy as compared to unmodified controls. This was particularly true of ANA modifications at or near the 3' end of the sense or antisense strands, while modification at the 5' end of the antisense strand resulted in complete loss of activity. Multiple ANA modifications within the sense strand were also well tolerated. Duplexes with ANA modifications at appropriate positions in both strands were generally more effective than duplexes with one modified and one unmodified strand. Initial evidence suggests that the loss of activity associated with ANA modification of the 5'-antisense strand may be due to reduced phosphorylation at this site by cellular kinases. Treatment of drug resistant cells with MDR1-targeted siRNAs resulted in reduction of P-glycoprotein (Pgp) expression, parallel reduction in MDR1 message levels, increased accumulation of the Pgp substrate rhodamine 123, and reduced resistance to anti-tumor drugs. Interestingly, the duration of action of some of the ANA-modified siRNAs was substantially greater than that of unmodified controls. These observations suggest that altritol modifications may be helpful in developing siRNAs with enhanced pharmacological effectiveness.
Figures



References
-
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
-
- Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. - PubMed
-
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. - PubMed
-
- Nawrot B, Sipa K. Chemical and structural diversity of siRNA molecules. Curr. Top. Med. Chem. 2006;6:913–925. - PubMed
-
- Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

