Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies
- PMID: 17267406
- PMCID: PMC1851655
- DOI: 10.1093/nar/gkm016
Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies
Abstract
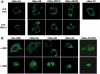
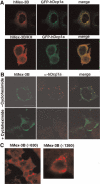
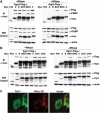
In Caenorhabditis elegans, the Mex-3 protein is a translational regulator that specifies the posterior blastomere identity in the early embryo and contributes to the maintenance of the germline totipotency. We have now identified a family of four homologous human Mex-3 genes, called hMex-3A to -3D that encode proteins containing two heterogeneous nuclear ribonucleoprotein K homology (KH) domains and one carboxy-terminal RING finger module. The hMex-3 are phosphoproteins that bind RNA through their KH domains and shuttle between the nucleus and the cytoplasm via the CRM1-dependent export pathway. Our analysis further revealed that hMex-3A and hMex-3B, but not hMex-3C, colocalize with both the hDcp1a decapping factor and Argonaute (Ago) proteins in processing bodies (P bodies), recently characterized as centers of mRNA turnover. Taken together, these findings indicate that hMex-3 proteins constitute a novel family of evolutionarily conserved RNA-binding proteins, differentially recruited to P bodies and potentially involved in post-transcriptional regulatory mechanisms.
Figures



References
-
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. - PubMed
-
- Labbe JC, Goldstein B. Embryonic development: a new SPN on cell fate specification. Curr. Biol. 2002;12:R396–398. - PubMed
-
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. - PubMed
-
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

