Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination
- PMID: 17267488
- PMCID: PMC1866129
- DOI: 10.1128/JVI.02589-06
Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination
Abstract
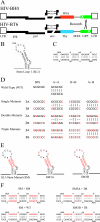
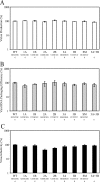
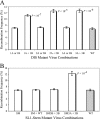
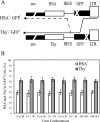
Frequent human immunodeficiency virus type 1 (HIV-1) recombination occurs during DNA synthesis when portions of the two copackaged RNAs are used as templates to generate a hybrid DNA copy. Therefore, the frequency of copackaging of genomic RNAs from two different viruses (heterozygous virion formation) affects the generation of genotypically different recombinants. We hypothesized that the selection of copackaged RNA partners is largely determined by Watson-Crick pairing at the dimer initiation signal (DIS), a 6-nucleotide palindromic sequence at the terminal loop of stem-loop 1 (SL1). To test our hypothesis, we examined whether heterozygous virion formation could be encouraged by manipulation of the DIS. Three pairs of viruses were generated with compensatory DIS mutations, designed so that perfect DIS base pairing could only occur between RNAs derived from different viruses, not between RNAs from the same virus. We observed that vector pairs with compensatory DIS mutations had an almost twofold increase in recombination rates compared with wild-type viruses. These data suggest that heterozygous virion formation was enhanced in viruses with compensatory DIS mutations (from 50% to more than 90% in some viral pairings). The role of the SL1 stem in heterozygous virion formation was also tested; our results indicated that the intermolecular base pairing of the stem sequences does not affect RNA partner selection. In summary, our results demonstrate that the Watson-Crick pairing of the DIS is a major determinant in the selection of the copackaged RNA partner, and altering the base pairing of the DIS can change the proportion of heterozygous viruses in a viral population. These results also strongly support the hypothesis that HIV-1 RNA dimers are formed prior to encapsidation.
Figures
Similar articles
-
Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging.J Virol. 2000 Jun;74(12):5729-35. doi: 10.1128/jvi.74.12.5729-5735.2000. J Virol. 2000. PMID: 10823883 Free PMC article.
-
HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization.J Mol Biol. 2007 Aug 24;371(4):1084-98. doi: 10.1016/j.jmb.2007.06.010. Epub 2007 Jun 9. J Mol Biol. 2007. PMID: 17599354
-
Conserved determinants of lentiviral genome dimerization.Retrovirology. 2015 Sep 29;12:83. doi: 10.1186/s12977-015-0209-x. Retrovirology. 2015. PMID: 26420212 Free PMC article.
-
Mechanisms and factors that influence high frequency retroviral recombination.Viruses. 2011 Sep;3(9):1650-1680. doi: 10.3390/v3091650. Epub 2011 Sep 9. Viruses. 2011. PMID: 21994801 Free PMC article. Review.
-
The retroviral RNA dimer linkage: different structures may reflect different roles.Retrovirology. 2004 Aug 18;1:22. doi: 10.1186/1742-4690-1-22. Retrovirology. 2004. PMID: 15317659 Free PMC article. Review.
Cited by
-
Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and gag sequences.RNA. 2016 Jun;22(6):905-19. doi: 10.1261/rna.055731.115. Epub 2016 Apr 19. RNA. 2016. PMID: 27095024 Free PMC article.
-
Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses.J Virol. 2010 Jul;84(13):6352-66. doi: 10.1128/JVI.00363-10. Epub 2010 Apr 14. J Virol. 2010. PMID: 20392841 Free PMC article.
-
HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein.Proc Natl Acad Sci U S A. 2016 Jan 12;113(2):E201-8. doi: 10.1073/pnas.1518572113. Epub 2015 Dec 28. Proc Natl Acad Sci U S A. 2016. PMID: 26712001 Free PMC article.
-
Mutations in matrix and SP1 repair the packaging specificity of a Human Immunodeficiency Virus Type 1 mutant by reducing the association of Gag with spliced viral RNA.Retrovirology. 2010 Sep 8;7:73. doi: 10.1186/1742-4690-7-73. Retrovirology. 2010. PMID: 20825656 Free PMC article.
-
Dimeric RNA recognition regulates HIV-1 genome packaging.PLoS Pathog. 2013 Mar;9(3):e1003249. doi: 10.1371/journal.ppat.1003249. Epub 2013 Mar 21. PLoS Pathog. 2013. PMID: 23555259 Free PMC article.
References
-
- Baba, S., K. Takahashi, S. Noguchi, H. Takaku, Y. Koyanagi, N. Yamamoto, and G. Kawai. 2005. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. (Tokyo) 138:583-592. - PubMed
-
- Bender, W., and N. Davidson. 1976. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7:595-607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

