mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes
- PMID: 17267594
- PMCID: PMC1892994
- DOI: 10.1073/pnas.0611125104
mRNA maturation by two-step trans-splicing/polyadenylation processing in trypanosomes
Abstract
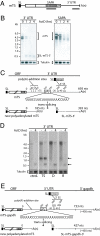
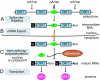
Trypanosomes are unique eukaryotic cells, in that they virtually lack mechanisms to control gene expression at the transcriptional level. These microorganisms mostly control protein synthesis by posttranscriptional regulation processes, like mRNA stabilization and degradation. Transcription in these cells is polycistronic. Tens to hundreds of protein-coding genes of unrelated function are arrayed in long clusters on the same DNA strand. Polycistrons are cotranscriptionally processed by trans-splicing at the 5' end and polyadenylation at the 3' end, generating monocistronic units ready for degradation or translation. In this work, we show that some trans-splicing/polyadenylation sites may be skipped during normal polycistronic processing. As a consequence, dicistronic units or monocistronic transcripts having long 3' UTRs are produced. Interestingly, these unspliced transcripts can be processed into mature mRNAs by the conventional trans-splicing/polyadenylation events leading to translation. To our knowledge, this is a previously undescribed mRNA maturation by trans-splicing uncoupled from transcription. We identified an RNA-recognition motif-type protein, homologous to the mammalian polypyrimidine tract-binding protein, interacting with one of the partially processed RNAs analyzed here that might be involved in exon skipping. We propose that splice-site skipping might be part of a posttranscriptional mechanism to regulate gene expression in trypanosomes, through the generation of premature nontranslatable RNA molecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Messenger RNA processing sites in Trypanosoma brucei.Mol Biochem Parasitol. 2005 Oct;143(2):125-34. doi: 10.1016/j.molbiopara.2005.05.008. Mol Biochem Parasitol. 2005. PMID: 15993496
-
Insights into the Regulation of mRNA Processing of Polycistronic Transcripts Mediated by DRBD4/PTB2, a Trypanosome Homolog of the Polypyrimidine Tract-Binding Protein.J Eukaryot Microbiol. 2016 Jul;63(4):440-52. doi: 10.1111/jeu.12288. Epub 2016 Jan 5. J Eukaryot Microbiol. 2016. PMID: 26663092
-
mRNA splicing in trypanosomes.Int J Med Microbiol. 2012 Oct;302(4-5):221-4. doi: 10.1016/j.ijmm.2012.07.004. Epub 2012 Sep 7. Int J Med Microbiol. 2012. PMID: 22964417 Review.
-
Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome.Future Microbiol. 2011 Apr;6(4):459-74. doi: 10.2217/fmb.11.20. Future Microbiol. 2011. PMID: 21526946 Review.
-
A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes.Genes Dev. 1994 Feb 15;8(4):491-501. doi: 10.1101/gad.8.4.491. Genes Dev. 1994. PMID: 7907303
Cited by
-
The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi.BMC Genomics. 2009 Aug 7;10:370. doi: 10.1186/1471-2164-10-370. BMC Genomics. 2009. PMID: 19664227 Free PMC article.
-
Proteomic analysis of Trypanosoma cruzi response to ionizing radiation stress.PLoS One. 2014 May 19;9(5):e97526. doi: 10.1371/journal.pone.0097526. eCollection 2014. PLoS One. 2014. PMID: 24842666 Free PMC article.
-
Regulation of transcription termination in the nematode Caenorhabditis elegans.Nucleic Acids Res. 2009 Nov;37(20):6723-36. doi: 10.1093/nar/gkp744. Epub 2009 Sep 9. Nucleic Acids Res. 2009. PMID: 19740764 Free PMC article.
-
Trypanosomes lack a canonical EJC but possess an UPF1 dependent NMD-like pathway.PLoS One. 2025 Mar 7;20(3):e0315659. doi: 10.1371/journal.pone.0315659. eCollection 2025. PLoS One. 2025. PMID: 40053537 Free PMC article.
-
Inhibition of mRNA maturation in trypanosomes causes the formation of novel foci at the nuclear periphery containing cytoplasmic regulators of mRNA fate.J Cell Sci. 2012 Jun 15;125(Pt 12):2896-909. doi: 10.1242/jcs.099275. Epub 2012 Feb 24. J Cell Sci. 2012. PMID: 22366449 Free PMC article.
References
-
- D'Orso I, De Gaudenzi JG, Frasch AC. Trends Parasitol. 2003;19:151–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

