Global reconstruction of the human metabolic network based on genomic and bibliomic data
- PMID: 17267599
- PMCID: PMC1794290
- DOI: 10.1073/pnas.0610772104
Global reconstruction of the human metabolic network based on genomic and bibliomic data
Abstract
Metabolism is a vital cellular process, and its malfunction is a major contributor to human disease. Metabolic networks are complex and highly interconnected, and thus systems-level computational approaches are required to elucidate and understand metabolic genotype-phenotype relationships. We have manually reconstructed the global human metabolic network based on Build 35 of the genome annotation and a comprehensive evaluation of >50 years of legacy data (i.e., bibliomic data). Herein we describe the reconstruction process and demonstrate how the resulting genome-scale (or global) network can be used (i) for the discovery of missing information, (ii) for the formulation of an in silico model, and (iii) as a structured context for analyzing high-throughput biological data sets. Our comprehensive evaluation of the literature revealed many gaps in the current understanding of human metabolism that require future experimental investigation. Mathematical analysis of network structure elucidated the implications of intracellular compartmentalization and the potential use of correlated reaction sets for alternative drug target identification. Integrated analysis of high-throughput data sets within the context of the reconstruction enabled a global assessment of functional metabolic states. These results highlight some of the applications enabled by the reconstructed human metabolic network. The establishment of this network represents an important step toward genome-scale human systems biology.
Conflict of interest statement
Conflict of interest statement: N.C.D., S.A.B., N.J., I.T., M.L.M., T.D.V., R.S., and B.Ø.P. and the University of California at San Diego disclose potential financial conflict of interest related to U.S. Patent Application (Pub. No. 20040029149), published on February 12, 2004.
Figures
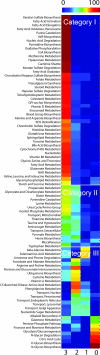
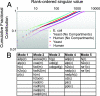

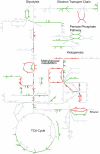
References
-
- International Human Gene Sequencing Consortium. Nature. 2004;431:931–945.
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. - PubMed
-
- McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, Sekhon M, Wylie K, Mardis ER, Wilson RK, et al. Nature. 2001;409:934–941. - PubMed
-
- Reed JL, Famili I, Thiele I, Palsson BO. Nat Rev Genet. 2006;7:130–141. - PubMed
-
- Price ND, Reed JL, Palsson B. Nat Rev Microbiol. 2004;2:886–897. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

