Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles
- PMID: 17267609
- PMCID: PMC1892985
- DOI: 10.1073/pnas.0608582104
Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles
Abstract
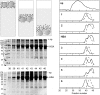
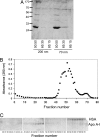
Due to their small size, nanoparticles have distinct properties compared with the bulk form of the same materials. These properties are rapidly revolutionizing many areas of medicine and technology. Despite the remarkable speed of development of nanoscience, relatively little is known about the interaction of nanoscale objects with living systems. In a biological fluid, proteins associate with nanoparticles, and the amount and presentation of the proteins on the surface of the particles leads to an in vivo response. Proteins compete for the nanoparticle "surface," leading to a protein "corona" that largely defines the biological identity of the particle. Thus, knowledge of rates, affinities, and stoichiometries of protein association with, and dissociation from, nanoparticles is important for understanding the nature of the particle surface seen by the functional machinery of cells. Here we develop approaches to study these parameters and apply them to plasma and simple model systems, albumin and fibrinogen. A series of copolymer nanoparticles are used with variation of size and composition (hydrophobicity). We show that isothermal titration calorimetry is suitable for studying the affinity and stoichiometry of protein binding to nanoparticles. We determine the rates of protein association and dissociation using surface plasmon resonance technology with nanoparticles that are thiol-linked to gold, and through size exclusion chromatography of protein-nanoparticle mixtures. This method is less perturbing than centrifugation, and is developed into a systematic methodology to isolate nanoparticle-associated proteins. The kinetic and equilibrium binding properties depend on protein identity as well as particle surface characteristics and size.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Probing the interactions of proteins and nanoparticles.Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2029-30. doi: 10.1073/pnas.0611610104. Epub 2007 Feb 6. Proc Natl Acad Sci U S A. 2007. PMID: 17284585 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

