Genome dynamics in a natural archaeal population
- PMID: 17267615
- PMCID: PMC1794283
- DOI: 10.1073/pnas.0604851104
Genome dynamics in a natural archaeal population
Abstract
Evolutionary processes that give rise to, and limit, diversification within strain populations can be deduced from the form and distribution of genomic heterogeneity. The extent of genomic change that distinguishes the acidophilic archaeon Ferroplasma acidarmanus fer1 from an environmental population of the same species from the same site, fer1(env), was determined by comparing the 1.94-megabase (Mb) genome sequence of the isolate with that reconstructed from 8 Mb of environmental sequence data. The fer1(env) composite sequence sampled approximately 92% of the isolate genome. Environmental sequence data were also analyzed to reveal genomic heterogeneity within the coexisting, coevolving fer1(env) population. Analyses revealed that transposase movement and the insertion and loss of blocks of novel genes of probable phage origin occur rapidly enough to give rise to heterogeneity in gene content within the local population. Because the environmental DNA was derived from many closely related individuals, it was possible to quantify gene sequence variability within the population. All but a few gene variants show evidence of strong purifying selection. Based on the small number of distinct sequence types and their distribution, we infer that the population is undergoing frequent genetic recombination, resulting in a mosaic genome pool that is shaped by selection. The larger genetic potential of the population relative to individuals within it and the combinatorial process that results in many closely related genome types may provide the basis for adaptation to environmental fluctuations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


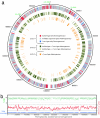

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

