Identification of novel functional TBP-binding sites and general factor repertoires
- PMID: 17268553
- PMCID: PMC1852848
- DOI: 10.1038/sj.emboj.7601550
Identification of novel functional TBP-binding sites and general factor repertoires
Abstract
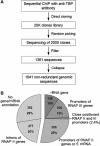
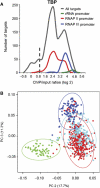
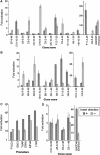
Our current knowledge of the general factor requirement in transcription by the three mammalian RNA polymerases is based on a small number of model promoters. Here, we present a comprehensive chromatin immunoprecipitation (ChIP)-on-chip analysis for 28 transcription factors on a large set of known and novel TATA-binding protein (TBP)-binding sites experimentally identified via ChIP cloning. A large fraction of identified TBP-binding sites is located in introns or lacks a gene/mRNA annotation and is found to direct transcription. Integrated analysis of the ChIP-on-chip data and functional studies revealed that TAF12 hitherto regarded as RNA polymerase II (RNAP II)-specific was found to be also involved in RNAP I transcription. Distinct profiles for general transcription factors and TAF-containing complexes were uncovered for RNAP II promoters located in CpG and non-CpG islands suggesting distinct transcription initiation pathways. Our study broadens the spectrum of general transcription factor function and uncovers a plethora of novel, functional TBP-binding sites in the human genome.
Figures




References
-
- Antequera F, Bird A (1994) Predicting the total number of human genes. Nat Genet 8: 114. - PubMed
-
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 - PubMed
-
- Berger SL (2002) Histone modifications in transcriptional regulation. Curr Opin Genet Dev 12: 142–148 - PubMed
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 - PubMed
-
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

