A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains
- PMID: 17270048
- PMCID: PMC1800850
- DOI: 10.1186/1471-2164-8-39
A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains
Abstract
Background: Noncoding RNA species play a diverse set of roles in the eukaryotic cell. While much recent attention has focused on smaller RNA species, larger noncoding transcripts are also thought to be highly abundant in mammalian cells. To search for large noncoding RNAs that might control gene expression or mRNA metabolism, we used Affymetrix expression arrays to identify polyadenylated RNA transcripts displaying nuclear enrichment.
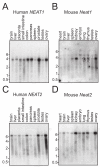
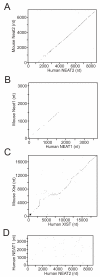
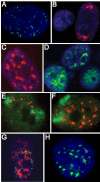
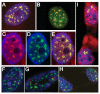
Results: This screen identified no more than three transcripts; XIST, and two unique noncoding nuclear enriched abundant transcripts (NEAT) RNAs strikingly located less than 70 kb apart on human chromosome 11: NEAT1, a noncoding RNA from the locus encoding for TncRNA, and NEAT2 (also known as MALAT-1). While the two NEAT transcripts share no significant homology with each other, each is conserved within the mammalian lineage, suggesting significant function for these noncoding RNAs. NEAT2 is extraordinarily well conserved for a noncoding RNA, more so than even XIST. Bioinformatic analyses of publicly available mouse transcriptome data support our findings from human cells as they confirm that the murine homologs of these noncoding RNAs are also nuclear enriched. RNA FISH analyses suggest that these noncoding RNAs function in mRNA metabolism as they demonstrate an intimate association of these RNA species with SC35 nuclear speckles in both human and mouse cells. These studies show that one of these transcripts, NEAT1 localizes to the periphery of such domains, whereas the neighboring transcript, NEAT2, is part of the long-sought polyadenylated component of nuclear speckles.
Conclusion: Our genome-wide screens in two mammalian species reveal no more than three abundant large non-coding polyadenylated RNAs in the nucleus; the canonical large noncoding RNA XIST and NEAT1 and NEAT2. The function of these noncoding RNAs in mRNA metabolism is suggested by their high levels of conservation and their intimate association with SC35 splicing domains in multiple mammalian species.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

