Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation
- PMID: 17273969
- PMCID: PMC1821101
- DOI: 10.1086/511888
Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation
Abstract
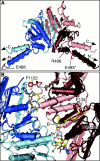
Mutations in the cohesin regulators NIPBL and ESCO2 are causative of the Cornelia de Lange syndrome (CdLS) and Roberts or SC phocomelia syndrome, respectively. Recently, mutations in the cohesin complex structural component SMC1A have been identified in two probands with features of CdLS. Here, we report the identification of a mutation in the gene encoding the complementary subunit of the cohesin heterodimer, SMC3, and 14 additional SMC1A mutations. All mutations are predicted to retain an open reading frame, and no truncating mutations were identified. Structural analysis of the mutant SMC3 and SMC1A proteins indicate that all are likely to produce functional cohesin complexes, but we posit that they may alter their chromosome binding dynamics. Our data indicate that SMC3 and SMC1A mutations (1) contribute to approximately 5% of cases of CdLS, (2) result in a consistently mild phenotype with absence of major structural anomalies typically associated with CdLS, and (3) in some instances, result in a phenotype that approaches that of apparently nonsyndromic mental retardation.
Figures



References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human SMC3 [accession number NM_005445] and human SMC1A [accession number NM_006306])
-
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/
-
- Human Genome Variation Society Mutation Nomenclature Recommendations, http://www.hgvs.org/mutnomen/
-
- NCBI Protein Database, http://www.ncbi.nlm.nih.gov/ (for struture coordinates for the Thermotoga SMC hinge domain [accession numbers 1GXLB and 1GXLA] and S. cerevisiae Smc1 head domain [accession numbers 1W1WA and 1W1WB])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CdLS) - PubMed
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

